4978
Quantifying the uncertainty of neural networks using Monte Carlo dropoutfor safer and more accurate deep learning based quantitative MRI
1Electrical and Electronics Engineering, Bogazici University, Istanbul, Turkey, 2Athinoula A. Martinos Center for Biomedical Imaging, Charlestown, MA, United States, 3Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom, 4Department of Radiology, Harvard Medical School, Boston, MA, United States, 5Department of Biomedical Engineering,College of Precision Instruments and Optoelectronics Engineering, Tianjin University, Tianjin, China
Synopsis
Neural networks reduce the data requirement for deep learning-based quantitative MRI, nonetheless their uncertainty/confidence has rarely been characterized. We implemented Monte Carlo dropout, a Bayesian approximation of Gaussian process, using U-Net that include dropout layers (active during training and inference) to address this. The uncertainty was calculated as the variance of predictions from 100 different dropout configurations. The estimates were calculated as the average of predictions. The proposed method also achieved higher accuracy in estimating FA and MD from only 3 diffusion-weighted images compared to standard U-Net, which was readily usable for other MRI applications (reconstruction, super-resolution, denoising, segmentation, classification).
Introduction
Quantitative MRI is an important and widely adopted tool to measure tissue properties. However, quantitative MRI often requires specialized sequences and many data samples from a lengthy scan for accurate and robust model fitting, which reduces its feasibility in practice. Emerging deep learning (DL) technologies using neural networks (NNs) have been employed to address this challenge, which have successfully generated high-quality quantitative metrics (e.g., T1, T2, mean diffusivity (MD), fractional anisotropy (FA)) using conventional contrast-weighted images and/or substantially reduced data1-5.Nonetheless, the uncertainty/confidence of these DL methods has rarely been characterized, which is as important as their accuracy. NNs may have varying uncertainty for estimating different quantitative metrics and/or in different regions/tissues. NNs may fail to generalize to test images that are not well represented by the training data due to the use of different hardware and imaging protocols and the presence of artifacts and structural pathology. It is therefore crucial to quantify the uncertainty/confidence of NNs for robustness characterization, risk management and potential human intervention on failure cases.
Monte Carlo dropout (MCDropout) provides an effective and feasible approach for uncertainty quantification. Dropout, which randomly switches off neurons in a NN during training (inactive during inference), is a useful regularizer to avoid overfitting. It was recently shown that using dropout also during inference can be interpreted as a Bayesian approximation of the Gaussian process6. Each dropout configuration, corresponding to a sub-network with slightly varying architecture, yields a different prediction as a sample of the approximate posterior distribution (Fig.1a). The uncertainty can be then quantified as the variance of predictions from many dropout configurations (e.g., 30~100). Moreover, the average of these predictions also achieves reduced uncertainty and higher accuracy (i.e., model averaging).
We propose to employ MCDropout for quantifying the uncertainty, preventing overfitting, and improving the performance of NNs for quantitative MRI. We demonstrate its effectiveness and feasibility by using slightly modified U-Net7, a NN commonly used in numerous MRI studies2,5,8,9, for synthesizing high-quality FA and MD values from one b=0 image and three diffusion-weighted image (DWI) volumes, an impossible task using DTI model fitting. (Code/data: https://anonymous.4open.science/r/dropout_ISMRM-4102/)
Methods
Data. Pre-processed diffusion data (1.25-mm isotropic, 18×b=0, 90×b=1000 s/mm2) of 42 subjects (10 for testing) from the Human Connectome Project (HCP)10-12 were used. For each subject, the first b=0 image and three DWI volumes along orthogonal directions were extracted. Ground-truth FA and MD volumes were derived from all 108 volumes using FSL’s “dtifit” function. The “aseg” brain segments from FreeSurfer were resampled to the diffusion image space.Training. A 3D U-Net was modified to include dropout layers (active during both training and inference) for decoding (Fig.1b, entitled “DU-Net”), which was essentially a standard U-Net if dropout rates were set to 0. Training was performed using the Tensorflow software with Adam optimizers on 64×64×64 image blocks. L1 loss was only computed within the brain parenchyma.
Evaluation. Experiments were performed with different numbers of subjects for training (1~32) and dropout rates (0~0.7). For each voxel, the uncertainty was calculated as the standard deviation over mean of 100 predictions, and the final estimate was calculated as the average of 1~100 predictions. The mean absolute errors (MAEs) between NN-estimated and ground-truth FA and MD values were calculated within the brain parenchyma and averaged for measuring accuracy.
Results
Exemplar FA predictions from two dropout configurations were slightly varying (Fig.2a-c) whereas the variance between two 100-prediction-averaged results was lower (Fig.2d-f). The uncertainty map for FA was spatially varying (Fig.2g,h) and tissue-dependent (Fig.2g,h,j). The uncertainty for FA was lower in the white matter than in cerebral and deep gray matter and was lowest in the corpus callosum. The uncertainty was generally lower for MD estimation, a simpler task, than for FA estimation.Figure 3 demonstrated the capability of MCDropout for detecting artifacts unseen in the training data (i.e., voxels within the letter “M” set to very bright or dark). Uncertainty maps clearly exhibited increased uncertainty within the letter “M”. The FA estimation was more sensitive to the voxel intensity increase, while MD estimation was more sensitive to voxel intensity decrease.
Dropout also increased the estimation accuracy (Fig.4). Even a single prediction from DU-Net outperformed that from a standard U-Net for FA and MD (1~4 training subjects) estimation. The average of 100 predictions from DU-Net achieved the highest accuracy. The improvement was most significant when the number of subjects for training was limited (1~4 subjects, 15%~25% lower MAE) since dropout avoided overfitting. Averaging predictions from DU-Net trained on data from one subject had almost equivalent performance compared to results from a standard U-Net trained on data from four subjects.
Figure 5 demonstrated that 0.1~0.2 dropout rates were optimal for achieving the highest accuracy, and averaging more predictions led to higher accuracy while performance plateaued after averaging 50 predictions.
Discussion and Conclusion
MCDropout provides an effective and feasible tool for safer and more accurate quantitative MRI, which does not change the architecture of NNs and the optimization or add training overhead. MCDropout can be easily used for any networks (variational/unrolled network) and tasks (reconstruction, denosing, superresolution, segmentation, classification) for MRI. Future work will quantify the uncertainty of NNs trained on data from healthy subjects on patient data.Acknowledgements
The T1w data at 0.7 mm isotropic resolution and diffusion data at 1.25 mm isotropic resolution were provided by the Human Connectome Project, WU-Minn-Ox Consortium (Principal Investigators: David Van Essen and Kamil Ugurbil; U54-MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University. This work was supported by the National Institutes of Health (grant numbers P41-EB015896, P41-EB030006, U01-EB026996, U01-EB025162, S10-RR023401, S10-RR019307, S10-RR023043, R01-EB028797, R03-EB031175, K99-AG073506), the NVidia Corporation for computing support, and the Athinoula A. Martinos Center for Biomedical Imaging.References
1. Golkov V, Dosovitskiy A, Sperl JI, et al. q-Space deep learning: twelve-fold shorter and model-free diffusion MRI scans. IEEE transactions on medical imaging. 2016;35(5):1344-1351.
2. Qiu S, Chen Y, Ma S, et al. Multiparametric mapping in the brain from conventional contrast‐weighted images using deep learning. Magnetic Resonance in Medicine. 2021.
3. Tian Q, Bilgic B, Fan Q, et al. DeepDTI: High-fidelity six-direction diffusion tensor imaging using deep learning. NeuroImage. 2020;219:117017.
4. Aliotta E, Nourzadeh H, Patel SH. Extracting diffusion tensor fractional anisotropy and mean diffusivity from 3‐direction DWI scans using deep learning. Magnetic Resonance in Medicine.
5. Sveinsson B, Chaudhari AS, Zhu B, et al. Synthesizing Quantitative T2 Maps in Right Lateral Knee Femoral Condyles from Multi-Contrast Anatomical Data with a Conditional GAN. Radiology: Artificial Intelligence. 2021:e200122.
6. Gal Y, Ghahramani Z. Dropout as a bayesian approximation: Representing model uncertainty in deep learning. Paper presented at: international conference on machine learning2016.
7. Falk T, Mai D, Bensch R, et al. U-Net: deep learning for cell counting, detection, and morphometry. Nature Methods. 2019;16(1):67-70.
8. Gong E, Pauly JM, Wintermark M, Zaharchuk G. Deep learning enables reduced gadolinium dose for contrast-enhanced brain MRI. Journal of Magnetic Resonance Imaging. 2018;48(2):330-340.
9. Chen KT, Gong E, de Carvalho Macruz FB, et al. Ultra–Low-Dose 18F-Florbetaben Amyloid PET Imaging Using Deep Learning with Multi-Contrast MRI Inputs. Radiology. 2018;290(3):649-656.
10. Sotiropoulos SN, Jbabdi S, Xu J, et al. Advances in diffusion MRI acquisition and processing in the Human Connectome Project. NeuroImage. 2013;80:125-143.
11. Glasser MF, Sotiropoulos SN, Wilson JA, et al. The minimal preprocessing pipelines for the Human Connectome Project. NeuroImage. 2013;80:105-124.
12. Glasser MF, Smith SM, Marcus DS, et al. The human connectome project's neuroimaging approach. Nature Neuroscience. 2016;19(9):1175-1187.
Figures
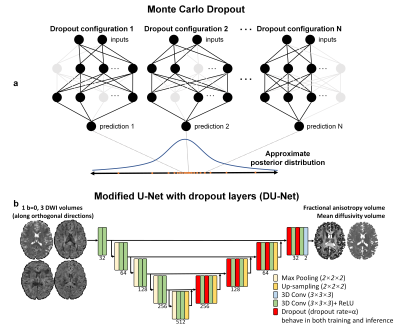
Figure 1. Monte Carlo dropout (MCDropout). MCDropout turns on dropout during inference and generates many varying predictions as samples from approximate posterior distribution of the Gaussian process for uncertainty quantification. The 3D U-Net (5 depth, 32 kernels at 1st depth) is modified to include dropout layers for decoding, which are active in both training and inference. The input includes 1 b=0 image volume and 3 diffusion-weighted image volumes along orthogonal directions. The output includes two volumes of fractional anisotropy and mean diffusivity values from DTI.
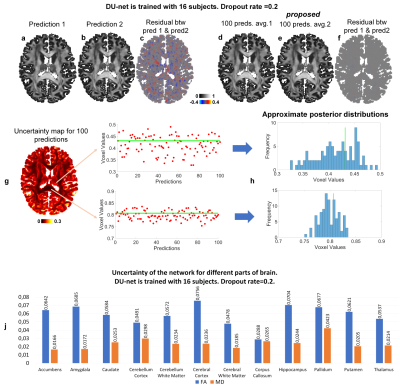
Figure 2. Uncertainty quantification. The difference between two FA predictions (a-c) and between two 100-prediction-averaged FA results (d-e). Uncertainty map (g) calculated from 100 predictions (ground-truth values indicated by green lines) and 100 predicated values of a single voxel from white matter with low uncertainty and a voxel from gray matter with high uncertainty, along with the approximate posterior distributions (histograms) derived from them are shown. The 10-subject mean values of the averaged uncertainty within different brain structures are listed (j).
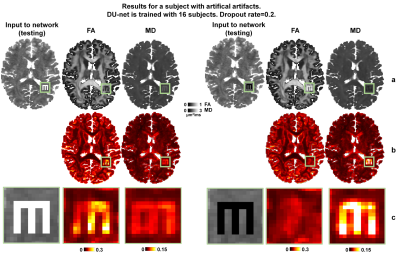
Figure 3. Uncertainty for image artifacts. Maps of FA and MD estimates (row a) and uncertainty (row b) obtained from 100 predictions on data with artificial image artifacts (i.e., voxels within the letter “M” set to very bright or dark).
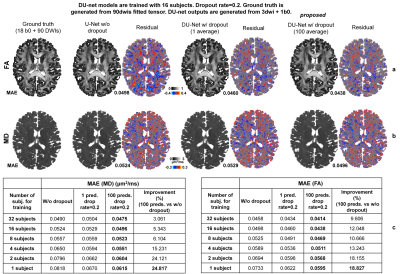
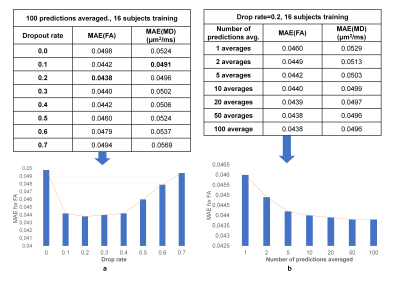
Figure 5. Optimal parameters for Monte Carlo dropout. The 10-subject means of the mean absolute error (MAE) between results and ground-truth values within the brain parenchyma for different dropout rates (ranging between 0.1 and 0.7) for the dropout layers of the DU-Net, and different numbers of predictions for averaging (ranging between 1 and 100) are listed.