4929
Accelerated Cardiac MR Imaging with Inline Cascaded Parallel Imaging and Generative Adversarial Neural Network (PI-GAN) Framework
Siyeop Yoon1, Xiaoying Cai1,2, Eiryu Sai1, Salah Assana1, Amine Amyar1, Kelvin Chow3, Amanda Paskavitz1, Julia Cirillo1, Warren J. Manning1, and Reza Nezafat1
1Department of Medicine, Cardiovascular Division, Beth Israel Deaconess Medical Center and Harvard Medical School, Boston, MA, United States, 2Siemens Medical Solutions USA, Inc., Boston, MA, United States, 3Siemens Medical Solutions USA, Inc., Chicago, IL, United States
1Department of Medicine, Cardiovascular Division, Beth Israel Deaconess Medical Center and Harvard Medical School, Boston, MA, United States, 2Siemens Medical Solutions USA, Inc., Boston, MA, United States, 3Siemens Medical Solutions USA, Inc., Chicago, IL, United States
Synopsis
We developed and evaluated an inline cascaded parallel imaging and generative adversarial network for cardiac MRI. The preliminary results show excellent image quality of the cardiac cine images in four heat-beat.
Introduction
Despite recent advances in compressed sensing (CS) and deep learning (DL) methods to accelerate MR imaging, there are still limitations for wide clinical adoption in cardiac MR imaging; therefore, parallel imaging remains the preferred image acceleration technique. CS reconstruction time remains long, even with the state-of-art hardware system, is only available for specific sequences (e.g., cardiac cine), and often uses spatial-temporal redundancy resulting in considerable temporal blurring. In addition, other than denoising algorithms, the majority of prior DL techniques have been tested offline using retrospective data without rigorous prospective evaluation. Furthermore, the generalizability of DL models for different sequences, slice orientation, and ease of integration into the standard clinical system remains challenging. In this study, we developed and evaluated an image reconstruction framework to accelerate cardiac MR imaging through a cascade of parallel imaging and generative adversarial neural network (PI-GAN) that combines widely available parallel imaging with GAN for the reconstruction of highly accelerated cardiac MR images. Images collected using late gadolinium enhancement (LGE) were used to train the PI-GAN model, which was later used to reconstruct cine images. Subsequently, we deployed the reconstruction model inline that allows the reconstruction of highly accelerated cardiac MR imaging on the scanner.Method
Figure 1.a shows schematic of the PI-GAN model. The fully-sampled k-space data are undersampled by collecting RGAN of ky lines, followed by RPI from undersampled k-space data divided into two regions: a) high-frequency and b) low-frequency. The resulting k-space data is first reconstructed using a matrix size, corresponding to a low-resolution parallel imaging image using conventional parallel imaging. In this study, we used a conventional GRAPPA acceleration factor of 2. The out of the initial PI reconstruction is de-aliased images with low spatial resolution. Subsequently, images reconstructed from GRAPPA are fed into a GAN reconstruction model to reconstruct the images into a larger matrix size. The architecture of the GAN consists of the following neural networks (Figure 1.b). 1) Generative Network has 23 dense residual blocks and 3 convolutional layers. This network removes artifacts and reconstructs high-resolution images from low-resolution images. 2) Discriminative network has 10 convolutional layers to train the generative network by discriminating whether the input of this network is the generated or original fully-sampled reconstructed. Furthermore, 3) Pre-trained VGG network extracts features and enables to compare the image at the feature level. Networks are trained using the following loss functions: 1) Pixel loss uses mean squared error of images. 2) Adversarial loss uses relative average GAN loss between predicted label distributions. 3) VGG feature based on L1-loss measures the difference between the VGG features. Furthermore, 4) Spatial frequency loss is L1-loss between the spatial frequencies of images. We set the weights of each cost function as adversarial loss = 0.005, pixel loss and spatial frequency loss = 0.01, and VGG loss = 1.0. The hyperparameters were batch size 36 and training patch size 48 by 48. We trained the neural networks for 210 epochs using Adam optimizer with learning rate= 0.0001, beta1=0.9, and beta2=0.999. We implemented the networks with Python and PyTorch library and trained on a DGX-1 workstation (NVIDIA, Santa Clara, California). The network was trained using 3D LGE images from 363 patients. Data were acquired at 3T (MAGNETOM Vida, Siemens Healthcare, Erlangen, Germany), and the k-space data were extracted from the vendor reconstruction pipeline. The datasets were divided into training (291 patients) and validation (72 patients). 3D LGE images were collected using RPI = 2. To simulate the RGAN rate 2-4 (i.e., overall acceleration between 4 and 8), we retrospectively discarded the phase-encoding k-space lines by selecting only 25% to 50% of the center of k-space lines. Afterwards, we generated training datasets consisting of pairs of GRAPPA-reconstructed images with low and high-resolution LGE images (Figure 2). We also integrated the GAN network inline using a Siemens prototype Framework for Image Reconstruction (FIRE)1. Low-resolution images were reconstructed in a Siemens Image Reconstruction Environment (ICE) and fed to PI-GAN using Fire in ISMRMRD format2. The reconstructed image is sent back to the ICE pipeline to correct distortion and generate a DICOM image. For evaluation, we prospectively acquired ECG-segmented cine bSSFP images from 29 subjects with GRAPPA rate 2 and PI-GAN accelerated segmented cine images and RGAN=4 (total acceleration of 8, 4 sec per slice). For DL-accelerated acquisition, we use the trained GAN for reconstruction. We quantified LVEF and LV mass from the cine images and compared the conventional and the proposed methods.Results
PI-GAN model, trained using LGE images, successfully reconstructed highly accelerated cine images in any slice orientation (Figure 3). Bland-Altman graph (Figure 4) shows excellent agreement between various structural and functional parameters measured using standard ECG-segmented cine images vs. PI-GAN accelerated cine images. Figure 5 shows inline reconstructed images on the scanner. The total reconstruction time for PI-GAN on the scanner was 133 seconds for 11 slices.Conclusion
PI-GAN can reconstruct highly accelerated cine images with different imaging contrast and orientation as the ones in the training set. An inline implementation allows seamless integration of the proposed acceleration approach into clinical workflow. Further studies are warranted to evaluate the performance of PI-GAN in other cardiac MR sequences.Acknowledgements
No acknowledgement found.References
1. Chow K, Kellman P, Xue H. "Prototyping Image Reconstruction and Analysis with FIRE". Society for Cardiovascular Magnetic Resonance 24th Annual Scientific Sessions (2021).
2. Inati, Souheil J., et al. "ISMRM Raw data format: A proposed standard for MRI raw datasets." Magnetic Resonance in Medicine 77.1 (2017): 411-421.
Figures
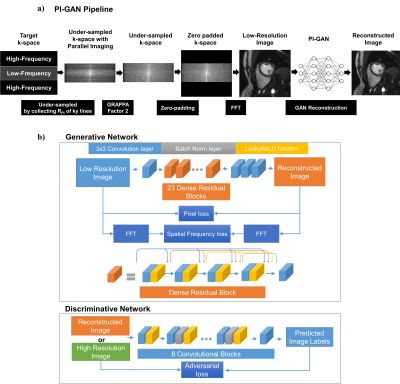
The schematic
of the PI-GAN model and architecture of GAN for image reconstruction. The low
spatial resolution from the initial PI reconstruction is fed into a GAN
reconstruction model to reconstruct the images into a high resolution.
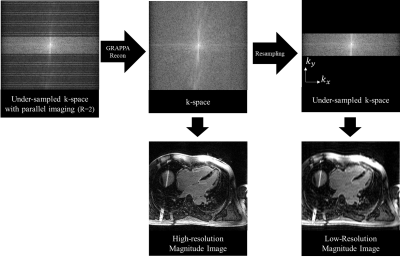
Data
preprocessing for synthesizing training and validation dataset. To generate a
low-resolution image retrospectively, the low-frequency region of the phase
encoding direction of the k-space is selected.
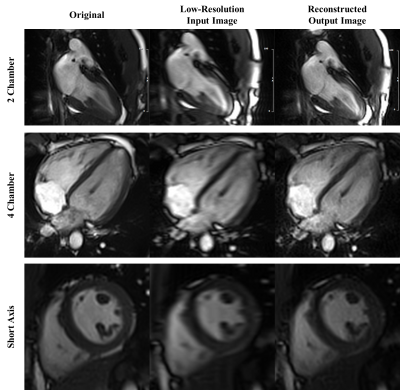
Example
images reconstructed using PI-GAN in 3 different slice orientations. The
proposed PI-GAN method was successful in recovering the anatomical features
with good blood-myocardium sharpness.
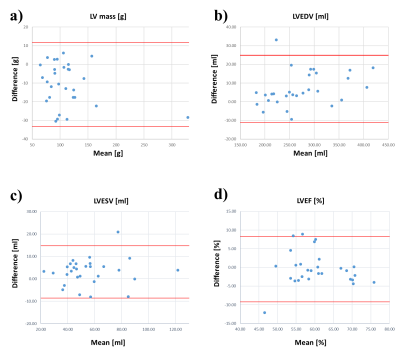
The
graphs show Bland-Altman analysis; the red line represents 95% limits of
agreement. LV mass-left ventricular mass; LVEDV-left ventricular end-diastolic
volume; LVESV-left ventricular end-systolic volume; LVEF-left ventricular
ejection fraction.
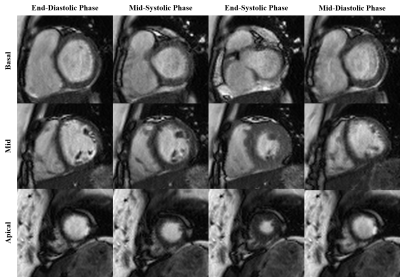
Inline reconstructed
cine images for different slices and cardiac phase acquired over four heartbeats
using PI-GAN with an acceleration factor of 8.
DOI: https://doi.org/10.58530/2022/4929