4928
Improved strain analysis of cine images by deep learning from DENSE: Comparison of a 3D Unet and an optical-flow net1Biomedical Engineering, University of Virginia, Charlottesville, VA, United States, 2University of Lyon, UJM-Saint-Etienne, INSA, CNRS UMR 5520, INSERM U1206, CREATIS, Saint-Etienne, France, 3Department of Radiology, University Hospital Saint-Etienne, Saint-Etienne, France, 4St. Francis Hospital, DeMatteis Center for Research and Education, Cardiac Imaging, Greenvale, NY, United States, 5Cardiovascular Magnetic Resonance Unit, The Royal Brompton Hospital, London, United Kingdom, 6National Heart and Lung Institute, Imperial College, London, United Kingdom, 7Department of Radiology and Imaging Sciences, Emory University School of Medicine, Atlanta, GA, United States, 8Department of Radiology, Stanford University, Stanford, CA, United States, 9Cardiovascular Division, Department of Medicine, University of Virginia Health System, Charlottesville, VA, United States, 10Radiology, University of Virginia, Charlottesville, VA, United States
Synopsis
Cine DENSE provides both myocardial contours and intramyocardial displacements. We propose to use DENSE to train deep networks to predict intramyocardial motion from contour motion. Two workflows were implemented: a two-step FlowNet2-based framework with a through-time correction network and a 3D (2D+t) Unet framework. Both networks depicted cardiac contraction and abnormal motion patterns. The 3D Unet showed excellent reliability for global circumferential strain (Ecc) and good reliability for segmental Ecc, and it outperformed commercial FT for both global and segmental Ecc.
Introduction
CMR myocardial strain imaging is used diagnostically and prognostically for many types of heart disease. Feature tracking (FT) is a widely used and convenient method for strain MRI, as it applies post-processing algorithms directly to standard cine images to assess strain. It is, however, less accurate than strain-dedicated acquisitions like displacement encoding with stimulated echoes (DENSE)1-4, especially for segmental strain. FT methods track myocardial contours rather than intramyocardial tissue because the myocardium presents uniform signal on cine MRI, lacking features to track. The intramyocardial motion is then (imperfectly) estimated using optical-flow based methods applied to the times series of endocardial and epicardial contours5. In contrast, DENSE directly measures intramyocardial tissue displacement; however, it requires additional acquisitions. As DENSE provides both myocardial contours and accurate intramyocardial tissue displacement information, we investigated the use of DENSE data to train deep networks to predict intramyocardial tissue motion from contour motion. This deep learning (DL) approach may provide the clinical convenience of FT and accuracy similar to DENSE.Methods
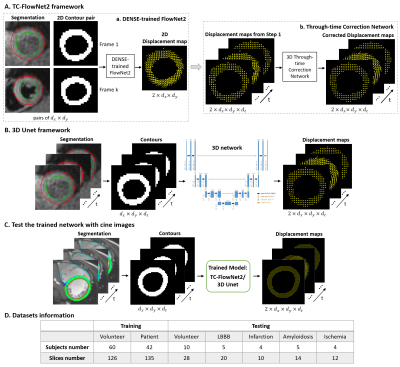
Two approaches were developed and evaluated: (a) a two-step FlowNet2-based framework with a through-time correction network (TC-FlowNet2), and (b) a 3D Unet. TC-FlowNet2 framework: This network was built upon a successful optical-flow convolutional neural network (CNN) called FlowNet26, which is widely used for frame-to-frame motion tracking of video imagery. We fine-tuned FlowNet2 using DENSE datasets, and we added a 3D through-time correction network to exploit the time dimension (Fig. 1A). 3D Unet framework: For this approach, a 3D Unet was trained to predict intramyocardial displacement from contour motion (Fig. 1B). For both approaches, during training, the inputs were a time series of myocardial contours derived from DENSE magnitude images and the ground truth data were DENSE tissue displacement measurements. Because DENSE and cine images at matched slice locations share similar motion patterns, we tested our trained model using contours derived from standard cine images (Fig. 1C). Data pre-processing for network training: We segmented the left-ventricular myocardium on DENSE and cine images, binarized the images by filling the myocardial area with 1 and the outside area and blood pool with 0, and cropped the images to a fixed size: Nx*Ny. Data augmentation was performed using 90° rotations. Cine images were scaled to match the resolution range of DENSE images. The input size for the FlowNet2-based network was two frames of endocardial and epicardial contours and the output of the DENSE-trained FlowNet2 was the frame-to-frame displacement field. The input of the through-time correction network was a stack of sequential displacements fields from DENSE-trained FlowNet2 with size of 2*Nx*Ny*Nt, where the factor of 2 accounts for displacements in two directions and Nt represents the number of temporal frames. The output was also size of 2*Nx*Ny*Nt. For the 3D Unet, the input size was Nx*Ny*Nt and the output size was 2*Nx*Ny*Nt. Datasets: Training datasets are described in Fig. 1D, and included a total of 60 volunteers and 42 patients with various pathologies such as left bundle branch block (LBBB), hypertrophic cardiomyopathy, dilated cardiomyopathy, coronary artery disease and hypertension. The model was tested on cine images of 10 volunteers and 18 patients using 3 short-axis views (base, mid-level and apex). For TC-FlowNet2, datasets were divided into two parts to separately train DENSE-trained FlowNet2 and the correction network, thus the testing dataset number (15 subject, 48 slices) was half the size as that used for the 3D Unet. Commercial feature-tracking (suiteHEART, Neosoft, WI) was also used to measure strain from cine images.Results
Fig. 2 shows examples comparing TC-FlowNet2, 3D Unet and DENSE for computing end-systolic displacement and circumferential strain (Ecc) for a healthy subject and a LBBB patient. In these examples, both methods detect cardiac contraction in the healthy volunteer and stretching of the septum in the LBBB patient, but TC-FlowNet2 shows less contraction. Fig. 3 shows examples comparing commercial FT, TC-FlowNet2, 3D Unet and DENSE for computing global and segmental circumferential strain-time curves, with the 3D Unet showing better agreement with the ground truth (DENSE). Correlation plots and Bland-Altman plots (Fig. 4A, B) show that 3D Unet outperformed both TC-FlowNet2 and commercial FT for global and segmental Ecc. Also, as shown in Table 1, the intraclass correlation coefficient (ICC), coefficient of variation (CoV), and Pearson correlation coefficient (Pearson CC) showed that the 3D Unet provides the best agreement with DENSE, where the 3D Unet achieved ICC = 0.89 for global Ecc and ICC = 0.75 for segmental Ecc. Although TC-FlowNet2 showed good linearity relationship with DENSE, it has a relatively big bias, leading to its high Pearson CC but relatively low ICC.Discussion and Conclusion
A 3D Unet, trained using DENSE datasets to predict intramyocardial motion from contour motion, outperformed both TC-FlowNet2 and commercial FT for the measurement of both global and segmental Ecc, for which DENSE data at matched locations served the reference standard.Acknowledgements
NIH R01HL147104, UVA Ivy Biomedical Innovation Fund and AHA 2020AHAPRE0000203801.References
1. Wehner GJ, Jing L, Haggerty CM, Suever JD, Chen J, Hamlet SM, Feindt JA, Mojsejenko WD, Fogel MA, Fornwalt BK. Comparison of left ventricular strains and torsion derived from feature tracking and DENSE CMR. Journal of Cardiovascular Magnetic Resonance. 2018 Dec;20(1):1-1.
2. Lin K, Meng L, Collins JD, Chowdhary V, Markl M, Carr JC. Reproducibility of cine displacement encoding with stimulated echoes (DENSE) in human subjects. Magn Reson Imaging. 2017;35:148-53.
3. Bilchick KC, Auger DA, Abdishektaei M, Mathew R, Sohn MW, Cai X, et al. CMR DENSE and the Seattle Heart Failure Model Inform Survival and Arrhythmia Risk After CRT. JACC Cardiovasc Imaging. 2020;13(4):924-36.
4. Mangion K, Loughrey CM, Auger DA, McComb C, Lee MM, Corcoran D, et al. Displacement Encoding With Stimulated Echoes Enables the Identification of Infarct Transmurality Early Postmyocardial Infarction. J Magn Reson Imaging. 2020;52(6):1722-31.
5. Schuster A, Hor KN, Kowallick JT, Beerbaum P, Kutty S. Cardiovascular Magnetic Resonance Myocardial Feature Tracking: Concepts and Clinical Applications. Circ Cardiovasc Imaging. 2016;9(4):e004077.
6. Ilg E, Mayer N, Saikia T, Keuper M, Dosovitskiy A, Brox T. Flownet 2.0: Evolution of optical flow estimation with deep networks. InProceedings of the IEEE conference on computer vision and pattern recognition 2017 (pp. 2462-2470).
Figures