4927
DL-Based LV Volume Segmentation to Compare Respiratory-Corrected and 4D XD-GRASP Respiratory Motion-Resolved Whole-Heart Reconstructions1Biomedical Engineering, Emory University/Georgia Institute of Technology, Atlanta, GA, United States, 2Biomedical Engineering, Georgia Institute of Technology, Atlanta, GA, United States, 3Siemens Medical Solutions USA, Princeton, NJ, United States, 4Diagnostic and Interventional Radiology, Lausanne University Hospital, Lausanne, Switzerland, 5Siemens Healthcare, Lausanne, Switzerland, 6Siemens Medical Solutions USA, Atlanta, GA, United States, 7Cardiology, Emory University School of Medicine, Atlanta, GA, United States, 8Radiology, Emory University School of Medicine, Atlanta, GA, United States
Synopsis
Rapid and accurate quantification of left ventricular volume is essential for studying cardiac function. In this work, two reconstruction techniques for free-breathing, ECG-gated, radial, golden-angle phyllotaxis acquisition of whole-heart CMR are assessed for their accuracy in quantifying left ventricular volume using deep learning (DL)-based automatic cardiac chamber segmentation. The off-line reconstruction that resolves 4D (3D+respiratory) images had better agreement between the DL-based segmentation and an expert’s manual segmentation than the in-line reconstruction that corrects for respiratory motion, as assessed by the average volume difference (p=0.026) and 3D Dice coefficients (p=0.032).
Introduction
In cardiovascular magnetic resonance studies, respiratory motion blurring and partial volume effects obscure the myocardial-blood border which in turn hinders the automated segmentation that is critical for rapid and accurate left ventricle (LV) volume and mass quantifications[1]. Current methods to compensate for respiratory motion include breath-holding for 2D imaging and free-breathing approaches for 3D whole-heart imaging, including navigator gating or correcting for motion using information from the acquired data[2, 3]. More recently, instead of correcting for respiratory motion, it was proposed that the motion extracted from the acquired data could be resolved and used to bin the acquired k-space data into different respiratory bins and perform reconstruction using compressed sensing (CS) (i.e., 4D XD-GRASP)[4]. This study uses a previously validated deep learning (DL)-based automatic segmentation algorithm[5] to assess the accuracy of LV volume segmentation in images reconstructed using: 1) in-line, motion-corrected method[3], and 2) off-line 4D XD-GRASP motion-resolved method[4]. We hypothesize that off-line 4D XD-GRASP reconstruction method would provide more accurate LV segmentation than motion-corrected method as assessed by the absolute volume difference (AVD) between the DL-based model and expert manual segmentation.Method
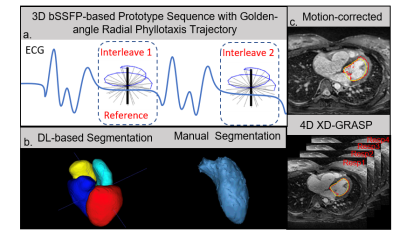
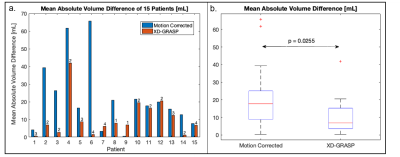
Data Acquisition: The study was performed on 15 patients at Emory University Hospital on a 1.5T scanner (MAGNETOM Avanto-fit, Siemens Healthcare, Erlangen, Germany). Data were acquired with a prototype ECG-gated 3D golden-angle radial phyllotaxis bSSFP sequence over 220mm3 FOV with an isotropic spatial resolution of 1.14mm3 (Fig.1a). Image Reconstruction: Respiratory motion-corrected images were reconstructed at the scanner by using each superior-inferior (SI) projection cross correlated with a reference SI projection to correct for relative respiratory motion[3]. 4D XD-GRASP reconstruction identifies a time-varying signal which lies within typical respiratory frequency ranges from a matrix of concatenated SI k-space projections. After motion extraction, k-space data were binned into four respiratory phases and CS was used to reconstruct each of the four highly under sampled respiratory phase image sets[4]. Image Segmentation: Per patient, one respiratory motion-corrected and four XD-GRASP respiratory motion-resolved whole-heart image volumes were processed using an image-to-image network that was trained on CT images and fine-tuned on multi-vendor MR images of variety of 3D MR sequences with a conditional variational autoencoder (I21-CVAE)[5] (Fig.1a, Left). The manual segmentation was done to segment LV by a trained expert in Mimics (Materialize, Inc) (Fig.1b, Right). The average volume difference (AVD) between manual and DL segmentation of each volume were computed and the minimum AVD of the four respiratory phases in 4D XD-GRASP was compared with motion-corrected method (Fig.2a). The 3D DICE coefficient was used to compare manual masks to the 4D XD GRASP masks and respiratory-corrected masks.Result
Four distinct respiratory phases were successfully reconstructed for 13 of 15 of the 4D XD-GRASP image datasets (Fig.1c). The smallest AVD in the respiratory motion-resolved 4D XD-GRASP reconstruction was lower than the respiratory motion-corrected reconstruction, 10.6±10.3 ml vs. 22.2±18.8 ml (mean ± standard deviation), p-value=0.026 (Fig. 2b.). The 3D DICE coefficient between the DL segmented masks and the manually segmented masks was higher for 4D XD-GRASP compared to the respiratory corrected images, 0.89±0.034 vs. 0.87±0.036, respectively (p-value=0.032).Discussion
This study shows that the automatic DL-based segmentations of LV using images reconstructed using off-line respiratory motion-resolved 4D XD-GRASP method are consistently closer to manually segmented masks than using images reconstructed with in-line respiratory motion-corrected as assessed by the difference between automatic and manual segmentation. The better segmentation using 4D XD-GRASP images likely results from the improved motion consistency and higher image quality, but a direct correlation between the two image sets needs to be further investigated.Conclusion
This study used a validated DL-based automated segmentation program as a metric to compare respiratory motion-corrected and motion-resolved reconstruction techniques. The results show the images generated from motion-resolved reconstruction technique provides an easier and more consistent quantification of LV volumes than the motion-corrected technique.Acknowledgements
We wish to acknowledge funding from the National Institute of Biomedical Imaging and Bioengineering, grant number R01-EB027774 (Oshinski); the Swiss National Science Foundation, grant number 173129 (Stuber)References
1. Kurkure U, Pednekar A, Muthupillai R, Flamm SD, Kakadiaris Ast IA. Localization and segmentation of left ventricle in cardiac cine-MR images. IEEE Trans Biomed Eng. May 2009;56(5):1360-70. doi:10.1109/tbme.2008.2005957
2. Bratis K, Henningsson M, Grigoratos C, et al. 'Image-navigated 3-dimensional late gadolinium enhancement cardiovascular magnetic resonance imaging: feasibility and initial clinical results'. J Cardiovasc Magn Reson. Dec 4 2017;19(1):97. doi:10.1186/s12968-017-0418-7
3.Piccini D, Littmann A, Nielles-Vallespin S, Zenge MO. Respiratory self-navigation for whole-heart bright-blood coronary MRI: methods for robust isolation and automatic segmentation of the blood pool. Magn Reson Med. Aug 2012;68(2):571-9. doi:10.1002/mrm.23247
4. Piccini D, Feng L, Bonanno G, et al. Four-dimensional respiratory motion-resolved whole heart coronary MR angiography. Magn Reson Med. Apr 2017;77(4):1473-1484. doi:10.1002/mrm.26221
5. Jacob A. et al., “Deep Learning-based 3D Whole Heart MR Segmentation with Limited Data”, SCMR 24th Annual Scientific Sessions, Feb 2021.
Figures