4902
Radial perfusion cardiac magnetic resonance imaging using deep learning image reconstruction.1Department of Medicine, Cardiovascular Division, Beth Israel Deaconess Medical Center, Boston, MA, United States
Synopsis
Myocardial perfusion assessment using cardiac MRI allows non-invasive assessment of myocardial ischemia. In myocardial perfusion sequence, imaging is collected after a saturation pulse. An alternative approach based on steady-state imaging with radial sampling has been recently proposed. However, image reconstruction using compressed sensing in steady-state myocardial perfusion remains long and clinically not feasible. In this study, we sought to develop a deep learning-based image reconstruction platform for myocardial perfusion imaging.
Purpose
First-pass myocardial cardiac MRI perfusion enables non-invasive assessment of coronary artery disease1. Clinically, perfusion images are collected using single-shot Cartesian sampling after a saturation pulse. Alternatively, free-running myocardial perfusion techniques are emerging using accelerated radial k-space sampling with golden-angle ordering. Such acceleration is needed to enable sufficient spatial resolution and slice coverage while maintaining a short acquisition window in the cardiac cycle. Compressed sensing has been key to achieving this acceleration, but reconstruction time remains long. In this study, we sought to develop a faster deep-learning-based approach for free-running myocardial perfusion imaging.Method
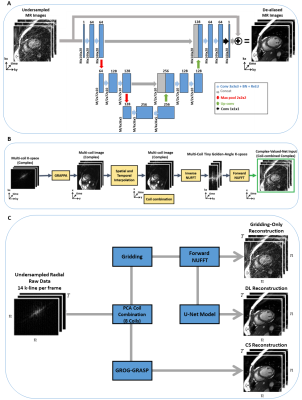
We implemented a 3D U-net architecture (2D+time), comprised of an encoder and a decoder with skip connections. The encoder consists of 6 convolutional layers with convolution filters of 3×3×3 followed by batch normalization and a max-pooling of 2×2×2 after every 2 convolutional layers. The number of feature maps was 64 in the first two layers, then increase to 128 in the next two layers and finally 256 in the last two layers. We use rectified linear activation function (ReLU)2 activation function. Each decoder level begins with a 3D convolutional transpose layer followed by a convolution. The up-sampled features are combined with the features from the corresponding level of the encoder part using concatenation. (Fig 1.A). We implemented and trained the network using PyTorch (Facebook, Menlo Park, California) on a DGX-1 workstation (NVIDIA Santa Clara, California, United States). Due to the limited availability of perfusion data, we trained our network using synthesized radial images from bSSFP cine Cartesian imaging. ECG-segmented k-space data of cine images from 503 patients (286 males, 55.4±15.8 years) were used to synthesize golden-angle radial k-space data and images (Fig 1.B) 3. The input of the network is the undersampled images and the output is the corresponding fully-sampled images. We concatenated real and imaginary components to enable real-valued processing of complex-valued data 2,4. The network was trained for 2,900 iterations using an L2 loss function, ADAM optimizer with 15% drop-out rate, batch size=16, and 20 consecutive time frames. Input/output images were normalized by the 95th percentile magnitude pixel intensity within the central region (i.e., 48×48) across 20 frames. The initial learning rate was 0.001, which was decreased by 5% every 100 iterations.To evaluate the model using perfusion images, we recruited 11 patients undergoing clinical cardiac MRI exams (MAGNETOM Vida, Siemens Healthcare, Germany). A free-running steady-state spoiled gradient recalled echo (SPGR) perfusion sequence was used to collect images during first-pass injection of 0.05 mmol/kg of gadobutrol with 10 mL of saline after contrast injection at a rate of 3 mL/s5. Data acquisition started 10-20s before the start of contrast injection and continued for ~2 min. Images were acquired with the following pulse sequence parameters: radial k-space sampling with golden angle ordering (111.25°)8 at 3 slices (apex, mid, and base), FOV=288×288 mm2, spatial resolution=2.0×2.0 mm2, slice thickness=8 mm, acquisition matrix = 144×144, TE=1.42 ms, and receiver bandwidth=1085 Hz/pixel.
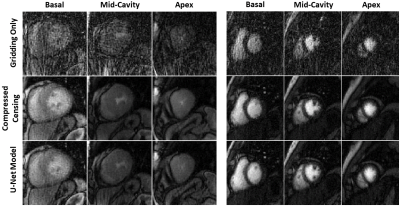
We reconstructed three slices (basal, mid, apical) for each subject using three methods: A) standard gridding, B) deep learning, and C) compressed censing using GROG-GRASP algorithm6. Each method began by first projecting 14 k-lines per phase and followed by principal component analysis (PCA) to produce 8 virtual coils. For standard gridding, we performed a forward NUFFT without further processing. The output of the NUFFT was input into the deep learning model which removed streaking artifacts. GROG-GRASP reconstruction was implemented in MATLAB (The MathWorks, Natick, MA).
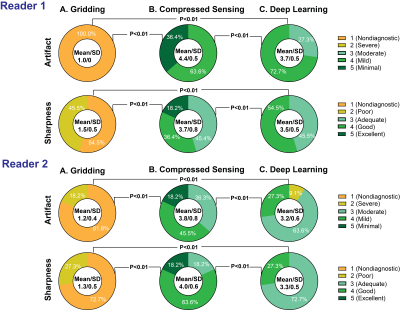
Image quality was evaluated by two independent readers for sharpness of wall enhancement and severity of streaking artifacts using a 5-point Likert scale (1=non-diagnostic to 5=minimal). The Wilcoxon Rank-Sum was used to assess statistical significance; a p-value < 0.05 was considered statistically significant.
Results
The deep learning model was able to significantly reduce the aliasing artifacts in the myocardial perfusion images, despite not being trained using any perfusion images (Fig. 2). Standard gridding reconstruction showed significant aliasing artifacts with poor average scores for artifacts (1.2±0.4) and sharpness (1.4±0.5) (Fig. 3). GROG-GRASP reconstruction provided better image quality compared to deep learning reconstruction for both artifact (4.1±0.7 vs 3.5±0.6, p < 0.01) and sharpness (3.9±0.7 vs 3.4±0.5 p < 0.01). However, all images were deemed to have moderate-to-minimal artifacts severity and sharpness scores with both deep learning and GROG-GRASP. The reconstruction time for ~159 phases of perfusion images was ~3.5 hours for compressed censing and ~60 seconds for the DL model.Conclusion
A deep learning based model, trained using cine images collected with Cartesian sampling, enables rapid reconstruction of myocardial perfusion images collected using a free-running myocardial perfusion sequence with accelerated radial k-space sampling.Acknowledgements
No acknowledgement found.References
1 Sirajuddin, A. et al. Ischemic Heart Disease: Noninvasive Imaging Techniques and Findings. RadioGraphics, 200125 (2021).
2 Ronneberger, O., Fischer, P. & Brox, T. in International Conference on Medical image computing and computer-assisted intervention. 234-241 (Springer).
3 Haji-Valizadeh, H. et al. Comparison of Complex k-Space Data and Magnitude-Only for Training of Deep Learning–Based Artifact Suppression for Real-Time Cine MRI. Front. Phys. 9: 684184. doi: 10.3389/fphy (2021).
4 Eo, T. et al. KIKI‐net: cross‐domain convolutional neural networks for reconstructing undersampled magnetic resonance images. Magnetic resonance in medicine 80, 2188-2201 (2018).
5 Haji‐Valizadeh, H. et al. Artifact reduction in free‐breathing, free‐running myocardial perfusion imaging with interleaved non‐selective RF excitations. Magnetic Resonance in Medicine 86, 954-963 (2021).
6 Benkert, T. et al. Optimization and validation of accelerated golden‐angle radial sparse MRI reconstruction with self‐calibrating GRAPPA operator gridding. Magnetic resonance in medicine 80, 286-293 (2018).
Figures