4895
Iterative Refinement of Expert Contours for Improved Ground-truth Quality in Intracranial Vessel Segmentation Neural Network Training1Bioengineering, University of California, Los Angeles, Los Angeles, CA, United States, 2Radiology, University of Southern California, Los Angeles, CA, United States, 3Radiation Oncology, University of California, Los Angeles, Los Angeles, CA, United States, 4Radiation Oncology, University of Southern California, Los Angeles, CA, United States, 5Biomedical Engineering, University of Southern California, Los Angeles, CA, United States
Synopsis
With a typical slice-by-slice labeling fashion, the manual contouring process is subject to large intra-observer variation, especially for small-sized intracranial arteries. We propose an iterative refinement approach for ground truth contours with the help of deep neural networks for intracranial lumen and vessel wall segmentations. We demonstrate that the approach improved the smoothness of the predicted contours and feature quantification, which can potentially boost the robustness of a neural network as a consequence of the reduced uncertainty in expert labels.
INTRODUCTION
When training deep neural networks for automatic vessel wall and lumen segmentation, manual labels from neuroradiologists are typically used as the ground truth. However, manually contouring the lumen and vessel wall can be challenged by inadequate signal contrast at vessel wall boundaries and is highly experience-dependent. Additionally, with a typical slice-by-slice labeling procedure, the manual contours of small intracranial vessels can exhibit artificial non-smoothness along the vessel direction, which may compromise the quality in downstream feature quantification, such as normalized wall index (NWI) 1. In this study, we propose an iterative refinement process to modify manual contours in ground-truth generation and investigate its effect on segmentation performance with deep learning models.METHODS
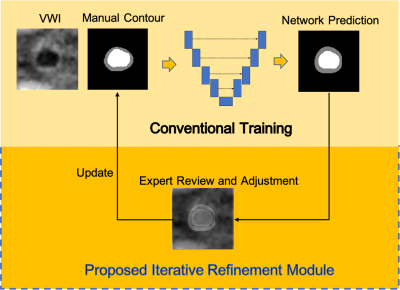
Data Preparation: Under IRB approval, we obtained magnetic resonance vessel wall imaging (MR VWI) from 80 patients with diagnosed ICAD. All imaging data were acquired on a 3-Tesla system (MAGNETOM Prisma; Siemens Healthcare, Erlangen, Germany) equipped with a 64-channel head/neck coil using a whole-brain VWI protocol 2. The following four vessel segments with a high likelihood of plaque presence were included for each patient: the intracranial internal carotid artery, the middle cerebral artery, the intracranial vertebral artery, and the basilar artery. Each segment contained 30 contiguous 2D cross-sectional slices with 0.55 mm slice thickness and 0.10 mm in-plane resolution generated in 3D Slicer 3. The manual lumen and vessel wall contours were labeled and refined by an experienced radiologist using ITK-SNAP 4.Refinement Process: The overall schema of the refinement process is illustrated in Fig. 1. The refinement process consists of three rounds of labeling and network training and starts with the initial labels generated by the radiologist based on VWI only. In the subsequent each round of manual contour refinement, the network predicted labels are overlayed with the corresponding VWI slices and are reviewed by the radiologist for either accepting the contours or applying necessary contour adaption. The refined contours are then used for the next round of network training and evaluation.
Segmentation Network Structure: A 2.5D UNet with ResNet backbone structure is adopted as the segmentation network, which takes three consecutive VWI slices, and outputs the labels of the background, lumen, and the vessel wall for the middle slice in three output channels 1. The base number of channels is 32, and each convolution block consists of two convolution layers, where a batch normalization layer is inserted after the first convolution layer. In each block the feature learned by the first convolution layer is added to the feature of the last convolution layer before going to a ReLU activation, to incorporate the previous information by skip-connections.
Evaluation of Network Segmentation Performance: We performed a five-fold cross-validation study, where each fold included 64 subjects for training and 16 subjects for testing. The network hyperparameters such as the number of training epochs, learning rate, and network depth, etc. were already optimized with respect to the validation results of our previous study and were kept fixed during the cross-validation process 1. Dice similarity coefficient (DSC) and mean surface distance (MSD) from the prediction to the ground truth were adopted as segmentation accuracy measures, across all folds of cross-validation. A clinically relevant measure -NWI, computed as the vessel wall area divided by the whole vessel area, was further incorporated as a measure to reflect the contour oscillations of a randomly selected vessel segment. DSC and MSD were reported for each network trained with the initial/revised contours and evaluated with the corresponding initial/revised contours.
RESULTS
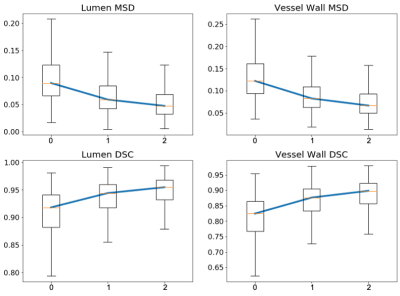
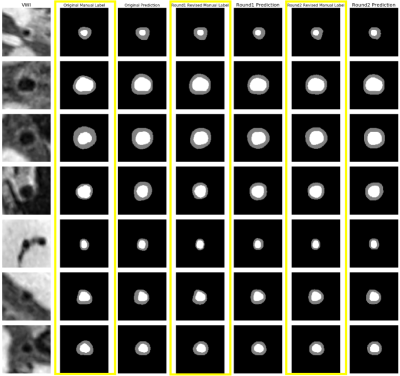
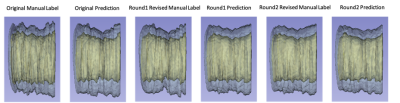
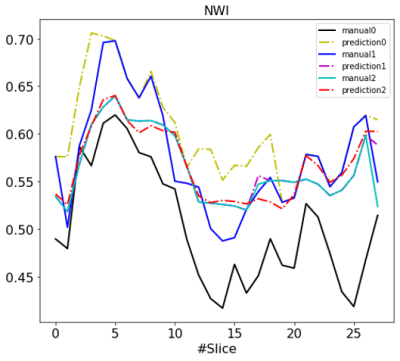
Fig. 2 reports the DSC and MSD of the lumen and the vessel wall classes vs. the number of refinement iterations in a box plot. The segmentation accuracy improved after the first and second iteration and tended to reach a plateau. Fig. 3 illustrates examples of the manual and network predicted segmentation in a progressive presenting fashion. The manual contour and network prediction more resembled each other as the number of iterations increased. Fig. 4 and Fig. 5 both demonstrate that the contours of the lumen and vessel wall both are smoother in the final refinement prediction compared to the initial network prediction trained with the initial manual labels.DISCUSSION
In this study, we demonstrated the value of iterative refining manual contours in mitigating the artificial non-smoothness associated with the current human labeling procedure. This approach also appears to improve the smoothness of the predicted contours as shown in Fig. 4 and in turn improves the smoothness of feature quantification as exampled by NWI in Fig. 5. A neural network trained through this strategy may be more robust as a consequence of reduced uncertainty in expert labeling.Acknowledgements
This work was supported in part by NIH/NHLBI R01 HL147355.References
1. Zhou H, Xiao J, Fan Z, Ruan D. Intracranial Vessel Wall Segmentation For Atherosclerotic Plaque Quantification. In: ; 2021:1416-1419. doi:10.1109/ISBI48211.2021.9434018
2. Yang Q, Deng Z, Bi X, et al. Whole-brain vessel wall MRI: A parameter tune-up solution to improve the scan efficiency of three-dimensional variable flip-angle turbo spin-echo. J Magn Reson Imaging. 2017;46(3):751-757. doi:10.1002/jmri.25611
3. Kikinis R, Pieper SD, Vosburgh KG. 3D Slicer: A Platform for Subject-Specific Image Analysis, Visualization, and Clinical Support. In: Intraoperative Imaging and Image-Guided Therapy. Springer New York; 2014:277-289. doi:10.1007/978-1-4614-7657-3_19
4. Yushkevich PA, Piven J, Hazlett HC, et al. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage. 2006;31(3):1116-1128. doi:10.1016/j.neuroimage.2006.01.015
Figures