4870
Accelerating 5D Whole-Heart MRA Reconstruction Using a Limited Number of Cardiac and Respiratory Frames1Biomedical Engineering, Emory University/Georgia Institute of Technology, Atlanta, GA, United States, 2Diagnostic and Interventional Radiology, Lausanne University Hospital, Lausanne, Switzerland, 3Siemens Healthcare, Lausanne, Switzerland, 4Lausanne University Hospital, Lausanne, Switzerland, 5Radiology, Emory University School of Medicine, Atlanta, GA, United States
Synopsis
Under-sampled reconstruction of retrospectively gated 5D free-running acquisition is time-consuming and computationally intensive. In this work, a limited reconstruction method is proposed which provides an efficient way to compute a static cardiac volume from free-breathing, ECG-free 5D free-running CMR acquisitions. Structural similarity index measure is used to compare the limited reconstructions with the fully reconstructed image at the physiologic state of interest. The proposed limited reconstruction method achieves a SSIM of 0.9 using only 22% of the full reconstruction time and an SSIM of 0.95 using 44% of the full reconstruction.
Introduction
Methods to compensate for physiologic motion in 3D whole-heart CMR include using synchronously measured respiratory and ECG signals, or using motion signals extracted from the acquired data[1-3]. The 5D free-running framework samples k-space continuously without gating and reconstructs images off-line by binning the raw data into distinct cardiac and respiratory phases according to the extracted motion signals[4]. Compressed sensing (CS) is used to reconstruct each highly under-sampled 3D k-space volume[5]. Such reconstruction is computationally expensive, as it requires an iterative algorithm to explore the sparsity across adjacent bins in both cardiac and respiratory dimensions. Some applications, such as coronary MRA, require reconstruction of only a single 3D volume and need a cost-efficient way to reconstruct data. Here, we propose to use a limited number of cardiac and respiratory bins surrounding the physiologic state of interest in CS reconstruction. We hypothesize that if we are only interested in reconstructing a single static 3D volume, using a subset of bins would create an acceptable final image and with significantly reduced computational cost.Method
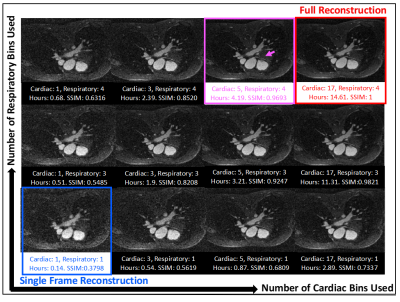
The study was performed using data on five pediatric patients who received Ferumoxytol (Fereheme, AMAG pharmaceutical) contrast at a concentration of 2-4 mg/kg on a 1.5T scanner (MAGNETOM Sola, Siemens Healthcare, Erlangen, Germany). A prototype, continuous, ungated, golden-angle, radial phyllotaxis[4], spoiled-GRE sequence was used (220mm3 FOV with 192 samples along each readout line, 1.1mm3 spatial resolution, 15° flip angle, TR=2.8/TE=1.6ms) to acquire 124,344 radial lines in 5.9 mins with 12 segments per interleave. The cardiac and respiratory signals were extracted and binned into four respiratory states and a variable number of cardiac phases such that each phase represented a 50ms window[4]. Alternating direction method of multiplier (ADMM)[6] was used to solve the reconstruction problem with sparsity regularization and data consistency. A full reconstruction using all binned data was performed to reconstruct a reference image volume at the cardiac resting phase during end-expiration, determined retrospectively by visual inspection of fully reconstructed data in cardiac and respiratory dimensions. A series of reconstructions were then done using the same raw data but with a variable number of total cardiac and respiratory bins centered at the resting cardiac phase (Fig.1). In total, there were 18 reconstructions (1,3,5,7,9, 12-20 cardiac bins, 1,3,4 respiratory bins) for each subject. Each limited reconstruction was compared against the full reconstruction by calculation of the 3D structural similarity index measure (SSIM)[7] of the image volume associated with the cardiac resting phase and end-expiration in both and plotted against the percent of bins used. The relative time used for each reconstruction was plotted against the relative bins used in the reconstruction.Result
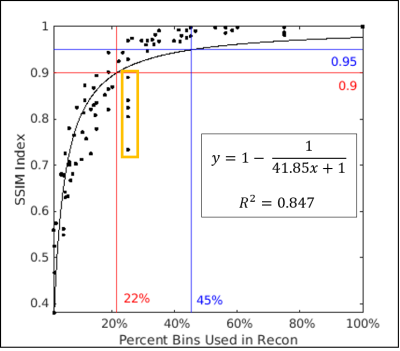
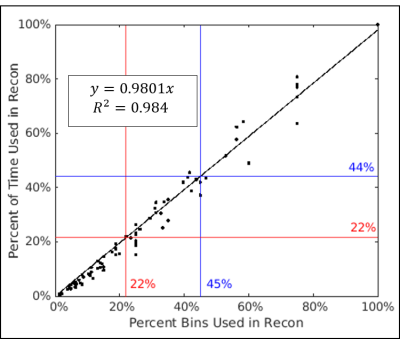
The full reconstruction time in five subjects ranged from 11.3 to 17 hours using all bins. An example subject is shown in Fig.1, where full reconstruction time equals to 14.6 hours using 68 bins (17 cardiac and 4 respiratory; red box, Fig.1). In the same subject, using 20 bins (5 cardiac and 4 respiratory), the image shows good agreement with fully reconstructed image with SSIM above 0.95 (magenta box in Fig.1). The reconstruction time is reduced to 4.2 hours and the right coronary artery can be visualized (magenta arrow). Using all five subjects’ data, a non-linear model of the form y = 1 – 1/ (ax+1) was fitted to the SSIM indices and percent of bins used in each limited reconstruction, where a=41.85 and the resulting coefficient of determination was 0.85 (Fig.2). Over all subjects, the intersection of this model with SSIM indices of 0.9 and 0.95 were at 22% and 45% of total bins, respectively. A strong linear relationship was also observed between the relative reconstruction time required and the ratio of bins used in each limited reconstruction (Fig.3).Discussion
We found that only 45% of total bins are needed to achieve an image quality that is functionally equivalent to the full reconstruction (SSIM of 0.95) and it required 44% of the total reconstruction time (6.5 ± 1.0 vs. 14.8 ± 2.2 hours, mean ± standard deviation). We also found that it is less optimal to use single respiratory phase in the limited reconstruction since even when all cardiac phases are used (25% bins, orange-boxed points, Fig.2), the reconstructed images still have greater structural distortions than those reconstructed using less bins.Conclusion
The computational cost of reconstructing images from a 5D free-running sequence for a single static phase was found to be directly proportional to the number of bins included in the calculation, which is non-linearly related to the improved SSIM indices. Only 22% of total reconstruction time is needed to achieve an image with SSIM of 0.9 (compared to the fully reconstructed data) and 44% of the time is needed to achieve SSIM of 0.95.Acknowledgements
We wish to acknowledge funding from the National Institute of Biomedical Imaging and Bioengineering, grant number R01-EB027774 (Oshinski); the Swiss National Science Foundation, grant number 173129 (Stuber)References
1. Coppo S, Piccini D, Bonanno G, et al. Free-running 4D whole-heart self-navigated golden angle MRI: Initial results. Magn Reson Med. Nov 2015;74(5):1306-16. doi:10.1002/mrm.25523
2. Piccini D, Littmann A, Nielles-Vallespin S, Zenge MO. Respiratory self-navigation for whole-heart bright-blood coronary MRI: methods for robust isolation and automatic segmentation of the blood pool. Magn Reson Med. Aug 2012;68(2):571-9. doi:10.1002/mrm.23247
3. Piccini D, Feng L, Bonanno G, et al. Four-dimensional respiratory motion-resolved whole heart coronary MR angiography. Magn Reson Med. Apr 2017;77(4):1473-1484. doi:10.1002/mrm.26221
4. Di Sopra L, Piccini D, Coppo S, Stuber M, Yerly J. An automated approach to fully self-gated free-running cardiac and respiratory motion-resolved 5D whole-heart MRI. Magnetic Resonance in Medicine. 2019;82(6):2118-2132. doi:https://doi.org/10.1002/mrm.27898
5. Feng L, Coppo S, Piccini D, et al. 5D whole-heart sparse MRI. Magn Reson Med. Feb 2018;79(2):826-838. doi:10.1002/mrm.26745
6. Boyd S, Parikh N, Chu E, Peleato B, Eckstein J. Distributed Optimization and Statistical Learning via the Alternating Direction Method of Multipliers. Found Trends Mach Learn. 2011;3(1):1–122. doi:10.1561/2200000016
7. Wang Z, Bovik A, Sheikh H, Simoncelli E. Image Quality Assessment: From Error Visibility to Structural Similarity. Image Processing, IEEE Transactions on. 05/01 2004;13:600-612. doi:10.1109/TIP.2003.819861
Figures