4864
AI-based Image Reconstruction of kt-accelerated Intracranial Dual-Venc 4D Flow MRI1Biomedical Engineering, Northwestern University, Chicago, IL, United States, 2Radiology, Northwestern University, Chicago, IL, United States, 3Neurology, Northwestern University, Chicago, IL, United States, 4Radiology, University of Greifswald, Greifswald, Germany
Synopsis
4D flow MRI provides a comprehensive assessment of hemodynamics through the 3D visualization and quantification of blood flow. However, its current clinical usage is hindered by long scan times. Recent developments have shown deep learning to be highly effective in accelerating image reconstruction, but no study has shown the its effectiveness in reconstructing highly undersampled cerebrovascular 4D flow MRI data. As such, we developed a CNN for the reconstruction of kt-accelerated intracranial dual-venc 4D flow MRI (R=5). We found that the CNN showed excellent SSIM values (0.94[0.93-0.95]) and moderate-to-good net flow and peak velocity agreement with the conventionally reconstructed data.
Introduction
4D flow MRI provides a comprehensive assessment of hemodynamics through the 3D visualization and quantification of blood flow. Recent studies have shown that quantitative analysis of intracranial blood flow is important for the understanding and management of a wide variety of diseases, such as intracranial atherosclerotic disease (ICAD) [1], intracranial aneurysms [2], and vascular malformations [3]. However, the current clinical usage of standard 4D flow MRI techniques is hindered by long scan times. Parallel Imaging has been an important contribution in accelerating the acquisition of MRI generally but is limited to certain acceleration factors and often suffers from long reconstruction times. Recent developments have shown deep learning to be highly effective in accelerating image reconstruction of highly undersampled MRI acquisitions [4-5], which maybe a basis to explore for 4D flow MRI. However, no study has shown the effectiveness of using deep learning in reconstructing highly undersampled cerebrovascular 4D flow MRI data. In this pilot study, we explored the feasibility of using a convolutional neural network (CNN) for the reconstruction of kt-accelerated intracranial dual-venc 4D flow MRI.Methods
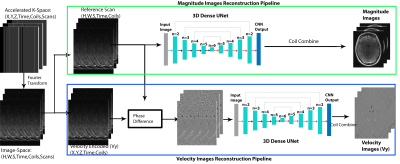
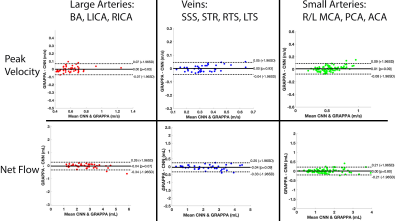
In this retrospective study, we used 12 intracranial 4D Flow MR scans from healthy control subjects (5 male, 7 female, 31.5±11.4 years) acquired on a 3T system (PRISMA, Siemens, spatial resolution = 0.98-1.13mm3, temporal resolution=82.6ms, venc=80/160cm/s). 10 of the subjects underwent two intracranial k-t accelerated GRAPPA, R=5 dual-venc 4D flow MRI scans on two separate days. In total, we acquired 22 scans from 12 subjects for CNN training and testing, where the standard k-t reconstruction (k-t GRAPPA, R=5) serves as ground truth data. For deep learning reconstruction, a hybrid 3D DenseNet/U-Net was employed as previously described [6]. Figure 1 provides a workflow for the CNN-reconstruction pipeline. After zero padding and Fourier transform, complex image-space datasets were used as CNN inputs. The real and imaginary components were separated and concatenated together for an input size (batch size=2,X=224,Y=192,slices,coils) and recombined (as real + j*imaginary) in the CNN output. Each time frame of the data was inputted separately into the reconstruction CNN. For 4D flow magnitude images, the reference scan was used as an CNN input, while the phase difference data (subtraction of velocity-encoded and reference scan) were used for flow images. The loss function consisted of a structural similarity index (SSIM) loss function + mean squared error. The reconstructed k-t GRAPPA with R=5 accelerated 4D flow data was used as ground-truth to assess performance of the CNN, and a 10-fold cross-validation method was used for training and testing.4D flow data analysis involved 3D flow visualization (streamlines) and peak velocity maximum intensity projections, MIPs (Figure 2). Additionally, net flow and peak velocity was calculated for each vessel using a custom-built Matlab tool [1]. In specific, centerlines were generated for all arterial branches of the circle of Willis and venous sinuses. Perpendicular 2D planes were placed along each centerline with 1mm distance. Net flow and peak velocity were obtained from each plane and the mean and standard deviation were reported for each COW artery (BA, LICA, RICA, L/R ACA, MCA, PCA), veins (SSS, STR, LTS, RTS)). The same planes and centerlines were used for both the CNN and conventionally reconstructed 4D flow MRI data. To assess the quality of the CNN reconstructed magnitude images, SSIM values (CNN vs conventional image reconstruction) were calculated and reported as median [interquartile range]. Bland-Altman comparisons were performed to compare the net flow and peak velocity between the AI-based reconstructed 4D flow and the conventional 4D flow in the large arteries, small arteries, and veins.
Results
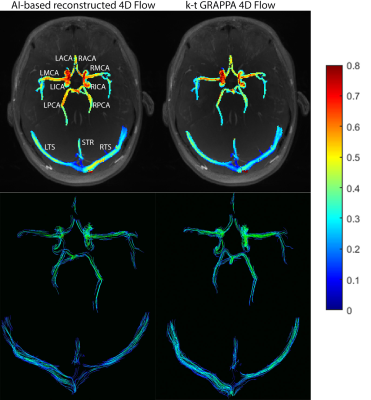
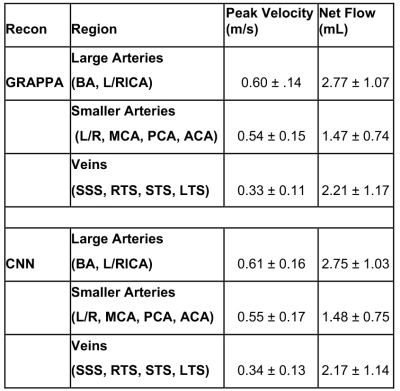
4D flow MRI reconstruction times for the CNNs was 129.6 ± 19.0 seconds, compared to ~600-900 seconds for k-t GRAPPA reconstruction. CNN based 4D flow reconstruction showed excellent SSIM values in the magnitude images, with a median SSIM of 0.94 [0.93-0.95]. Figure 2 provides examples of the systolic velocity MIPs and 3D streamlines for conventional 4D flow compared to the CNN-reconstructed data. CNN reconstructed cerebrovascular 4D flow MRI showed similar flow patterns and velocity mappings as conventional 4D flow.Figure 3 provides the Bland-Altman plots across the entire cohort at the large arteries (BA, LICA, RCA), smaller arteries (L/R MCA, PCA, ACA), and veins (SSS, LTS, RTS, STR). Additionally, Table 1 provides the flow and velocity measures for both the CNN reconstructed and conventional 4D flow. Across all comparisons, the CNN showed low bias and moderate to good agreement with the conventionally reconstructed data, showing 11-15.2% difference from the mean conventional 4D flow values.
Discussion
We demonstrated the feasibility of utilizing a CNN for the reconstruction of cerebrovascular dual-venc 4D flow MRI, showing moderate to good net flow and peak velocity agreement with the conventionally reconstructed 4D flow MRI. A future direction of this study is to explore higher 4D flow MRI acceleration factors with AI-based reconstruction in order to reduce the 4D flow scan time without sacrificing hemodynamic accuracy.Acknowledgements
This work was funded by the National Institute of Health (NIH 1R01HL149787, NIH 1R21NS122511)References
1. Vali, A., et al., Semi-automated analysis of 4D flow MRI to assess the hemodynamic impact of intracranial atherosclerotic disease. Magn Reson Med, 2019. 82(2): p. 749-762.
2. Amili, O., et al., Hemodynamics in a giant intracranial aneurysm characterized by in vitro 4D flow MRI. PLoS One, 2018. 13(1): p. e0188323.
3. Aristova, M., et al., Standardized Evaluation of Cerebral Arteriovenous Malformations Using Flow Distribution Network Graphs and Dual-venc 4D Flow MRI. J Magn Reson Imaging, 2019.
4. Haji-Valizadeh, H., et al., Highly accelerated free-breathing real-time phase contrast cardiovascular MRI via complex-difference deep learning. Magn Reson Med, 2021. 86(2): p. 804-819.
5. Vishnevskiy, V., J. Walheim, and S. Kozerke, Deep Variational Network for Rapid 4D Flow MRI Reconstruction. Nature Machine Intelligence, 2020. 2.
6. Berhane, H., et al., Fully automated 3D aortic segmentation of 4D flow MRI for hemodynamic analysis using deep learning. Magn Reson Med, 2020. 84(4): p. 2204-2218.
Figures