4837
Surface-Volume Registration Produces Atlases More Realistic than Either Surface or Volume Registration1Department of Radiology and Biomedical Research Imaging Center (BRIC), The University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
Synopsis
We show that spatial registration by simultaneously considering both tissue contrast and cortical geometry produces brain atlases that are more reflective of individuals than registration by considering only one of the two factors alone. Registration with tissue contrast alone produces atlases with blurred cortical structures. Registration with cortical geometry alone produces surface atlases that are artificially sharp but do not reflect surface attributes of individuals.
Introduction
Brain atlases should ideally capture key structural details of a population of individuals. Existing brain atlases often fall short of this goal by focusing on preserving one type of structural detail but at the expense of another. More specifically, conventional registration methods typically consider either volumetric tissue contrast or cortical surface geometry, but not both at the same time. Volumetric registration alone blurs cortical structures as it does not explicitly consider cortical geometry. Cortical surface registration alone neglects the spatial contextual information provided by the tissues, often resulting in implausible subcortical displacements. Here, we show that surface-volume registration harnessing both cortical geometry and volumetric tissue contrast produces volume and surface brain atlases that are more reflective of individuals.Materials and Methods
The dataset consisted of structural MR scans of 37 subjects aged between 2 weeks and 24 months, recruited as part of the UNC/UMN Baby Connectome Project1 (BCP). We preprocessed the data using our infant-centric pipeline2-4, generating skull-stripped T1-weighted (T1w) images, tissue segmentation maps, and white and pial cortical surfaces. We evaluated three spatial alignment methods for atlas construction: (i) Spherical Demons5 (SD) for cortical surface registration; (ii) symmetric normalization6 (SyN) for volumetric registration; and (iii) surface-constrained dynamic elasticity model7 (SCDEM) for surface-volume registration. We employed SD to generate atlases of cortical attributes in a spherical space and SyN and SCDEM to generate both cortical surface and volumetric atlases.We constructed the atlases at each month with data of subjects scanned within the time interval $$$\mathcal{W}_{\tau}=[(\tau-\delta \tau),(\tau+\delta \tau)]$$$, where $$$(\tau, \delta \tau) \in\{(0.5,1.5),(1,1.5),(2,1.5),\ldots,(24,1.5)\}$$$ (unit: months). SD-derived spherical atlases of surface attributes (i.e., mean curvature and average convexity) were obtained by aligning the spherical maps of white surfaces to iteratively updated average maps of geometric attributes. The cortical surface atlases were given by the final average maps of mean curvature and average convexity at the end of groupwise registration. To build volumetric atlases, we first constructed the surface-volume atlas at 12 months. This was accomplished by groupwise registration (via SyN and SCDEM) of the tissue segmentation maps of subjects included in $$$\mathcal{W}_{12}$$$. We used tissue segmentation maps instead of intensity images to account for the dynamically evolving tissue contrast in infant brain MRI. At the end of volumetric groupwise registration, we fused the warped tissue segmentation maps using majority voting and averaged the warped T1w images and the white and pial cortical surfaces using temporal kernel regression. The volume and surface atlases were subsequently warped to other time points from 2 weeks to 24 months using parallel transported longitudinal deformations8 estimated using SyN and SCDEM.
Results
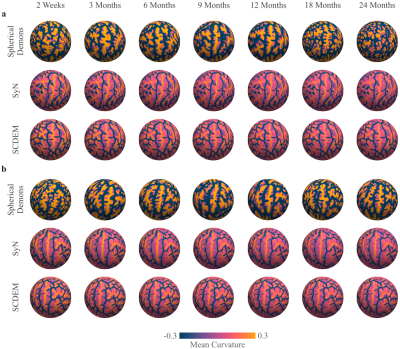
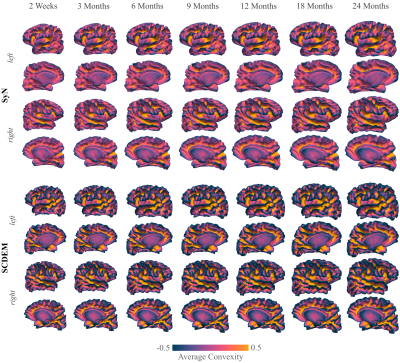
We show the mean curvature maps of the cortical atlases generated with SD, SyN, and SCDEM in Fig. 1; only surface-volume registration preserves the geometric features of the cortical surfaces, resulting in detailed secondary and tertiary folding patterns captured by the mean curvature of the surface atlases at all time points. SD and SyN based mean curvature maps are inconsistent across the two-year span and exhibit uncharacteristic sulcal and gyral arrangement with altered folding patterns.Comparing the white surface atlases given by SyN and SCDEM (Fig. 2), we can observe that primary cortical folds are captured by SCDEM only, as indicated by the sharp average convexity maps. SyN-based cortical atlases exhibit less cortical details with atypical folding pattern.
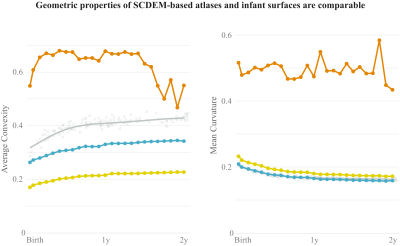
Quantitative analysis reveals that SCDEM yields mean curvature and average convexity curves that are comparable to generalized additive mixture models (GAMMs) of mean curvature and average convexity measurements of the individuals (Fig. 3). Note that the atlases gives in general lower values due to the inevitable smoothing of small folds during atlas construction.
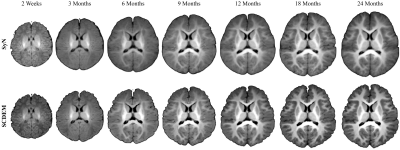
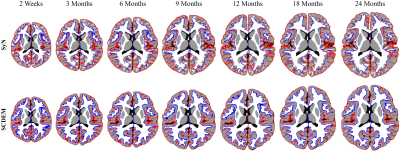
SCDEM generates volumetric T1w atlases with rich anatomical details (Fig. 4) that align well with the cortical surface atlases at tissue interfaces (Fig. 5). In contrast, SyN blurs anatomical details and does not promote spatial consistency between the cortical surface and volumetric atlases.
Conclusion
Considering both cortical geometry and volumetric tissue contrast for spatial alignment in atlas construction results in atlases that are more representative of individuals both in terms of volumetric anatomical details and cortical surface attributes.Acknowledgements
This work was supported in part by the United States National Institutes of Health (NIH) through grant EB008374.References
1. Howell BR, et al. The UNC/UMN Baby Connectome Project (BCP): An overview of the study design and protocol development. NeuroImage. 2019;185:891 - 905.
2. Wang L, et al. Volume-based analysis of 6-month-old infant brain MRI for autism biomarker identification and early diagnosis. International Conference on Medical Image Computing and Computer-Assisted Intervention. 2018;411 - 419.
3. Li G, et al. Measuring the dynamic longitudinal cortex development in infants by reconstruction of temporally consistent cortical surfaces. NeuroImage. 2014;90:266 - 279.
4. Li G, et al. Construction of 4D high-definition cortical surface atlases of infants: Methods and applications. Medical Image Analysis. 2015;25:22 - 36.
5. Yeo BTT, et al. Spherical Demons: Fast diffeomorphic landmark-free surface registration. IEEE Transactions on Medical Imaging. 2010;29(3):650 - 668.
6. Avants BB, et al. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Medical Image Analysis. 2008;12(1):26 - 41.
7. Ahmad S, et al. Surface-constrained volumetric registration for the early developing brain. Medical Image Analysis. 2019;58:101540.
8. Ahmad S, et al. Surface-volume consistent construction of longitudinal atlases for the early developing brain. International Conference on Medical Image Computing and Computer-Assisted Intervention. 2019;815-822.
Figures