4817
Improving Across-Dataset Brain Tissue Segmentation Using Transformer1Department of Biomedical Engineering, Columbia University, New York, NY, United States, 2Department of Applied Mathematics, Columbia University, New York, NY, United States, 3Department of Psychiatry, Columbia University, New York, NY, United States
Synopsis
Despite achieving compelling performance, many deep learning automated brain tissue segmentation solutions struggle to generalize to new datasets due to properties inherent to MRI scans. We propose TABS, a new transformer-based deep learning architecture that achieves state-of-the-art-performance, generalization, and consistency. We tested TABS on three datasets of differing field strands and acquisition parameters. TABS outperformed RAUnet on our performance testing and remained consistent across test-retest repeated scans from a separate dataset. Moreover, TABS achieved impressive generality performance and even improved in performance across datasets. We believe TABS represents a generalized and accurate brain tissue segmentation alternative.
Introduction
Brain tissue segmentation involves classifying voxels of the brain as white matter (WM), gray matter (GM), or cerebrospinal fluid (CSF); this process has demonstrated great clinical utility by highlighting subtle structural changes within the brain. Alzheimer’s1, Parkinson’s2, Huntington’s3, and Multiple Sclerosis4 among many other neurological conditions5 are linked with distinct volumetric changes more easily detected following brain tissue segmentation. However, manual segmentation of brain tissue is extremely labor intensive and difficult even for experts.Various automated segmentation algorithms have been proposed to circumvent these issues, ranging from traditional thresholding and Gaussian-Mixture-Model (GMM)6,7 based approaches to more recently deep Convolutional Neural Networks (CNNs)8-12. Automated segmentation has proven to be challenging due to properties inherent to the MRI scans themselves including noise, artifacts, and low contrast between tissues13. As such, there remains a high demand for accurate, generalized, and robust brain tissue segmentation tools.
Transformers have recently emerged as a compelling alternative to CNNs, and transformer-based implementations outperform CNNs on many applications including segmentation14. In this study, we introduce Transformer-based Automated Brain Segmentation (TABS), a new transformer-based deep learning (DL) architecture that achieves state-of-the-art brain tissue segmentation performance and generalization.
Methods
The architecture of our proposed model is shown in Figure 1a. TABS consists of a 5-layered CNN encoder with squeeze-excitation blocks15 before each down-sampling operation. The encoder portion is followed by a transformer module, which then feeds back into a CNN decoder. We compared TABS to RAUnet, a Unet variant known to outperform the traditional Unet architecture for segmentation16. Both models were trained using ground truths generated from FAST6 and optimized using Adam with mean-squared-error loss. Each model took a 3D MRI input (192x192x192) and outputted a three channel probability map (3x192x192x192) corresponding to each tissue type.We performed three experiments evaluating model performance, generality, and consistency. Our experimental pipeline is visualized in Figure 1b. Firstly, we collected MRI scans of healthy participants from three datasets: DLBS17, SALD18, and IXI19, following the pre-processing protocol outlined in Feng et al20. We split each into 3:1:1 train/val/test groups with homogenous age distributions between each group. The acquisition parameters and characteristics for each dataset are outlined in Figure 2. Both TABS and RAUnet were trained on each of the datasets for 350 epochs, with early stopping based on validation performance. They were also trained on the pooled total dataset for 200 epochs following the same protocol. All of the models were tested on the dataset they were trained on. Afterwards, the pre-trained models were applied across datasets to evaluate generality. Specifically, models trained on 3T SALD/DLBS scans were evaluated on 1.5T IXI scans, and models trained on 3T SALD/DLBS scans were evaluated on one another. Finally, to evaluate model consistency, we compared the segmentation outputs from FAST, RAUnet, and TABS on paired test-retest scans from the COBRE dataset. We quantified probability-based segmentation performance using Pearson/Spearman correlations and mean-squared-error, and we evaluated map-based segmentation performance using DICE and Jaccard Index.
Github Repository for TABS can be found at: https://github.com/raovish6/TABS
Results
For the model performance testing, TABS outperformed RAUnet on the majority of the datasets. TABS achieved better probability-based performance across all datasets and brain tissue types. Additionally, TABS attained better DICE scores on the DLBS and total datasets as well as for GM and CSF on the SALD dataset. These results are summarized in Figure 3a. Qualitative evaluation of the segmentation outputs suggests that TABS is more reliable as well shown in Figure 3b.We also found that TABS could generalize across datasets exceptionally well. Interestingly, TABS performance appeared to increase when applied across dataset; TABS pre-trained on DLBS and SALD performed better on IXI than TABS trained on IXI itself. RAUnet failed to generalize well in comparison. These results are outlined in Figure 4.
Finally, our test-retest results indicate that TABS is significantly more consistent than the RAUnet model and of comparable consistency to FAST. These results are shown in Figure 5.
Discussion
This study investigated the performance of a new transformer-based brain tissue segmentation model. TABS showcased superior performance compared to the prior state-of-the-art Unet variant. Additionally, TABS also demonstrated impressive generalization and consistency performance, atypical of traditional CNN models.Given the prevalence of brain tissue segmentation as a clinical screening and pre-processing tool, it is not practical to retrain a DL model every time a scan is obtained using different acquisition parameters. While DL implementations until now have achieved compelling results when trained on one dataset, it is unclear whether they properly generalize. The three datasets used in this study significantly varied with the purpose of emulating extreme scenarios where input scans differ. TABS has proven exceptionally reliable despite these obstacles, even improving across datasets. Our results indicate that TABS’ performance could be dependent on the quality of the ground truths, performing better when trained on 3T scans. Therefore, we believe our pre-trained TABS model could perform well on lower quality scans that would otherwise be difficult to segment.
Conclusion
In this study, we propose TABS, a new transformer-based DL model achieving state-of-the-art brain tissue segmentation performance, generality, and reliability. Notably, we achieve impressive generalization, even improving across datasets of different field strands and acquisition parameters.Acknowledgements
This work was supported by and performed at Zuckerman Mind Brain Behavior Institute MRI Platform, a shared resource, and Columbia MR Research Center site.References
Ewers M, Sperling RA, Klunk WE, et al. Neuroimaging markers for the prediction and early diagnosis of Alzheimer's disease dementia. Trends in Neurosciences. 2011; 34(8):430–442.
Hutchinson M, Raff U. Structural Changes of the Substantia Nigra in Parkinson’s Disease as Revealed by MR Imaging. AJNR Am. J. Neuroradiol. 2000; 21(4):697-701.
Ciarmiello A, Cannella M, Lastoria S, et al. Brain White-Matter Volume Loss and Glucose Hypometabolism Precede the Clinical Symptoms of Huntington's Disease. J. Nucl. Med. 2006; 47(2):215-222.
Rovaris M, Filippi M. Magnetic resonance techniques to monitor disease evolution and treatment trial outcomes in multiple sclerosis. Curr. Opin. Neurol. 1999; 12(3):337-344.
Smith-Bindman R, Miglioretti DL, Johnson E, et al. Use of Diagnostic Imaging Studies and Associated Radiation Exposure for Patients Enrolled in Large Integrated Health Care Systems, 1996-2010. JAMA. 2012; 307(22):2400–2409.
Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden markov random field model and the expectation-maximization algorithm. IEEE Trans. Med. Imag. 2001; 20(1):45–57.
Dora L, Agrawal S, Panda R, et al. State-of-the-Art Methods for Brain Tissue Segmentation: A Review. IEEE Rev. Biomed. Eng. 2017; 10:235-249.
Bernal J, Kushibar K, Cabezas M, et al. Quantitative Analysis of Patch-Based Fully Convolutional Neural Networks for Tissue Segmentation on Brain Magnetic Resonance Imaging. IEEE Access. 2019; 7:89986-90002
Zhang F, Breger A, Cho KI, et al. Deep Learning based segmentation of brain tissue from diffusion MRI. NeuroImage. 2021; 233:117934.
Lee B, Yamanakkanavar N, Choi JY. Automatic segmentation of brain MRI using a novel patch-wise U-net Deep Architecture. PLOS ONE. 2020; 15(8):e0236493.
Yamanakkanavar N, Lee B. Using a Patch-Wise M-Net Convolutional Neural Network for Tissue Segmentation in Brain MRI Images. IEEE Access. 2020; 8:120946-120958.
Akkus Z, Galimzianova A, Hoogi A. et al. Deep Learning for Brain MRI Segmentation: State of the Art and Future Directions. J Digit Imaging. 2017; 30:449–459.
Scherrer B, Forbes FC, Garbay C, et al. Distributed Local MRF Models for Tissue and Structure Brain Segmentation. IEEE Trans. Med. Imag. 2009; 28(8):1278-1295.
Wang W, Chen C, Ding M, et al. TransBTS: Multimodal Brain Tumor Segmentation Using Transformer. arXiv. 2021; 2103.04430.
Hu J, Shen L, and Sun G. Squeeze-and-Excitation Networks. CVPR. 2018; 7132-7141.
Ni Z, Bian G, Zhou X, et al. RAUNet: Residual Attention U-Net for Semantic Segmentation of Cataract Surgical Instruments. ICONIP. 2019.
Rodrigue KM, Kennedy KM, Devous MD, et al. β-Amyloid burden in healthy aging: regional distribution and cognitive consequences. Neurology. 2012; 78(6):387-95
Wei D, Zhuang K, Ai L, et al. Structural and functional brain scans from the cross-sectional Southwest University adult lifespan dataset. Sci Data. 2018; 5:180134.
Ixi Dataset. Brain Development. (n.d.). Retrieved from https://brain-development.org/ixi-dataset/.
Feng X, Lipton ZC, Yang J, et al. Estimating brain age based on a uniform healthy population with deep learning and structural magnetic resonance imaging. Neurobiol. Aging. 2020; 91:15-25
Vaswani A, Shazeer N, Parmar N, et al. Attention Is All You Need. NeurIPS. 2017; 30
Figures

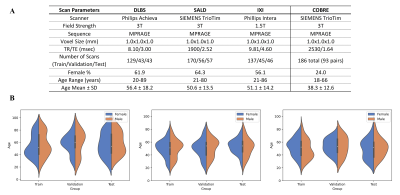
A. Acquisition and scanner parameters for each of the three primary datasets evaluated and the test-retest COBRE dataset. Each of the three primary datasets were collected using different scanners, with DLBS/SALD collected using 3T field strands while IXI was collected using 1.5T field strands. B. Visualized age distributions for the train/val/test splits for each of the datasets. Data was split to ensure homogenous age distributions.
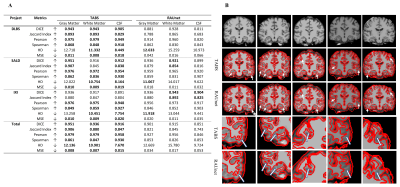
A. Performance results for models trained and tested on the same dataset. Metrics were evaluated for each tissue type and compared across TABS and RAUnet as indicated by the boldings. Arrow signs indicate metric directionality. B. Qualitative evaluation of TABS versus RAUnet outputs. The top row consists of the medial axial MRI slide. The following rows display the overlaid WM segmentation output for TABS and RAUnet respectively. Arrows indicate areas where TABS succeeded as opposed to RAUnet.
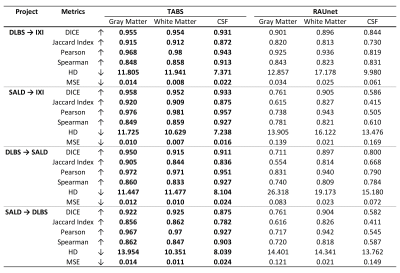
Performance results for models trained on one dataset and tested on another. The first two rows indicate performance for models trained on 3T DLBS/SALD and tested on 1.5T IXI. The last two columns display performance for the models trained on 3T DLBS/SALD tested on one another. Metrics were evaluated for each tissue type and compared across TABS and RAUnet as indicated by the boldings. Arrow signs indicate metric directionality.
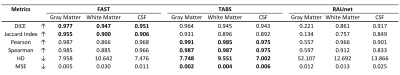
Test-retest results for FAST, TABS, and RAUnet tested on repeated scans from the COBRE dataset. We expected the more consistent models to display greater similarity between segmentation outputs for the repeated scans. Metrics were evaluated for each tissue type and compared across TABS, RAUnet, and FAST as indicated by the boldings. Arrow signs indicate metric directionality.