4816
Motion-robust dynamic abdominal MRI using k-t GRASP and dynamic dual-channel training of super-resolution U-Net (DDoS-UNet)1Biomedical Magnetic Resonance, Otto von Guericke University, Magdeburg, Germany, 2Institute for Medical Engineering, Otto von Guericke University, Magdeburg, Germany, 3Research Campus STIMULATE, Otto von Guericke University, Magdeburg, Germany, 4Data and Knowledge Engineering Group, Otto von Guericke University, Magdeburg, Germany, 5Faculty of Computer Science, Otto von Guericke University, Magdeburg, Germany, 6German Center for Neurodegenerative Disease, Magdeburg, Germany, 7Center for Behavioral Brain Sciences, Magdeburg, Germany, 8Leibniz Institute for Neurobiology, Magdeburg, Germany
Synopsis
Cartesian sampling techniques are available to speed up the measurement of dynamic MRI, such as k-t GRAPPA. However, radial samplings, such as iGRASP, are more robust to motion and can be applied for abdominal dynamic MRI. In this work, k-t GRAPPA inspired iGRASP has been created (so-called k-t GRASP)–which acquires the subsequent time points by starting the initial spoke of the time point with an angle with the last spoke of the previous time point, and extended by super-resolution reconstruction of dynamic abdominal MRI using DDoS-UNet. The method was evaluated in 3D dynamic data of four subjects with retrospective undersampling.
Introduction
For dynamic imaging of abdominal MRI, it is often necessary to speed up the image acquisition for assessing physiological movements. Several techniques have been developed for image acquisition acceleration. The DDoS-UNet [1], which employed the dynamic dual-channel training for super-resolution reconstruction, illustrated the ability to reconstruct dynamic MRI from highly undersampled data (the highest demonstrated undersampling level is 6.25% of k-space). In addition, techniques exploiting the redundant data in the k-t domain, such as k-t GRAPPA [2], have demonstrated the potential of achieving fast dynamic imaging. On the other hand, radial sampling techniques, such as iGRASP [3], have shown their potential of being inherently more motion-robust than Cartesian sampling schemes, and can be employed for fast dynamic MRI. HoweveThus, in this work, a combination of the aforementioned techniques into the k-t GRASP technique has been developed and used to perform super-resolution of dynamic abdominal MRIs using DDoS-UNet for highly undersampled data.Methods
k-t GRASP SchemeInspired by k-t GRAPPA and iGRASP, the proposed k-t GRASP technique continuously samples spokes with the golden angle (111.25°) over timepoints. For a given slice, the first spoke of the initial time point (TP1) is acquired at 0°, then the subsequent spokes of the same time point are acquired with a continuous angle increment of 111.25°. The first spoke of the second time point (TP2) is acquired with an angle increment of 111.25° with respect to the last spoke of TP1. In this manner, the angles are not only continuous within each time point but also over adjacent time points. Such sampling is performed in-plane, and they are then stacked together over the slice dimension to obtain the stack-of-stars in 3D. Fig. 1 illustrates the comparison between iGRASP and k-t GRASP. The authors hypothesize that for a dual-channel approach like DDoS-UNet, which encodes the spatial as well as the temporal information, such continuous supply of the missing spokes will assist in better reconstruction quality.
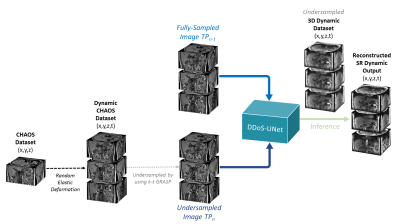
DDoS-UNet - Network Training and Implementation
Fig. 2 shows the network overview. The DDoS-UNet model comprises dual-channel inputs of an undersampled image of the current time point (TPn) and the corresponding fully-sampled image of the earlier time point (TPn-1). Therefore, the CHAOS challenge dataset (T1 dual images, in-, and opposed-phase) [4] was used to generate the dynamic data artificially using random elastic deformation [5] for dual-channel training. This data was then undersampled in-plane [6] using the kt-GRASP scheme with different spokes. 3D abdominal dynamic MRIs of four subjects were used for testing. Loss during training was minimized using the perceptual loss network with L1 loss and was also minimized using Adam optimizer for eight epochs. The DDoS-UNet performs patch-based super-resolution with 3D patches with the patch size of 243. For training, a stride of 6 was selected in the slice dimension and 12 for the others. For testing, a stride of 3 was chosen with the same patch size for all dimensions. The implementation was done using PyTorch.
Data Acquisition and Undersampling
The abdominal image acquisition was performed on a 3T Siemens Magnetom Skyra with the scanning parameters: GRE, T1w Flash 3D, Fat Saturation, TR 2.27-2.4ms, TE 0.97-1.02ms, voxel size 0.91x0.91x4-0.99x0.99x4.0mm. The acquired data was considered as fully-sampled and artificially undersampled with different levels of undersampling in this research: 10%, 6.25%, and 4% of the k-space data of the fully-sampled data, considering the fully-sampled as the number of spokes equals to twice the matrix size of the acquisition. For example, an image with 256x256 matrix size has a fully-sampled of 512 spokes. Therefore, 10%, 6.25%, and 4% undersamplings have 51, 32, and 20 spokes, respectively.
Results
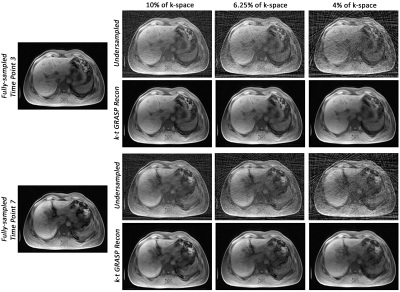
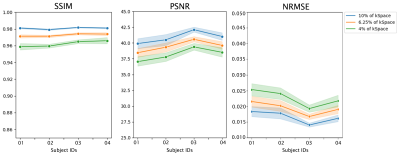
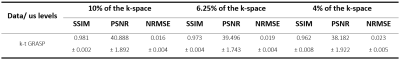
Fig. 3 portrays an example of the reconstructed images using k-t GRASP and DDoS-UNet of different undersampling levels and its ground-truth. The quantitative analysis was measured using structural similarity index (SSIM), peak signal-to-noise ratio (PSNR), and normalized root mean square error (NRMSE). Fig. 4 illustrates the SSIM, PSNR and NRMSE of each subject for different undersampling levels. Moreover, table 1 shows the average value of SSIM, PSNR, and NRMSE along with the standard deviation of all subjects for all time points against its ground-truth. It can be seen that k-t GRASP with DDoS-UNet can reconstruct the images from highly undersampled data. It can be observed that for 4% of the k-space, even though the network managed to achieve an average SSIM of 0.962 (as can be seen in Table 1), image smoothing becomes evident.Conclusion
This research presents k-t GRASP combined with DDoS-UNet, for performing super-resolution reconstruction of highly undersampled dynamic abdominal MRIs. The approach has shown high similarity to the fully-sampled ground-truth images: both quantitatively and qualitatively. This can be further extended by incorporating the sampling information directly with the reconstruction process of DDoS-UNet.Acknowledgements
This work was conducted within the context of the International Graduate School MEMoRIAL at Otto von Guericke University (OVGU) Magdeburg, Germany, kindly supported by the European Structural and Investment Funds (ESF) under the programme “Sachsen-Anhalt WISSENSCHAFT Internationalisierung“ (project no. ZS/2016/08/80646).References
[1] Sarasaen C, Chatterjee S, Nürnberger A, Speck O. DDoS: Dynamic Dual-Channel U-Net for Improving Deep Learning Based Super-Resolution of Abdominal Dynamic MRI. Magn Reson Mater Phy. 2021;34:S44. DOI: http://doi.org/10.1007/s10334-021-00947-8.
[2] Huang F, Akao J, Vijayakumar S, Duensing GR, Limkeman M. k‐t GRAPPA: A k‐space implementation for dynamic MRI with high reduction factor. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine. 2005 Nov;54(5):1172-84.
[3] Feng L, Grimm R, Block KT, Chandarana H, Kim S, Xu J, Axel L, Sodickson DK, Otazo R. Golden‐angle radial sparse parallel MRI: combination of compressed sensing, parallel imaging, and golden‐angle radial sampling for fast and flexible dynamic volumetric MRI. Magnetic resonance in medicine. 2014 Sep;72(3):707-17.
[4] Kavur AE, Gezer NS, Barış M, Aslan S, Conze PH, Groza V, Pham DD, Chatterjee S, Ernst P, Özkan S, Baydar B. CHAOS challenge-combined (CT-MR) healthy abdominal organ segmentation. Medical Image Analysis. 2021 Apr 1;69:101950.
[5] Pérez-García F, Sparks R, Ourselin S. TorchIO: a Python library for efficient loading, preprocessing, augmentation and patch-based sampling of medical images in deep learning. Computer Methods and Programs in Biomedicine. 2021 Jun 17:106236.
[6] Soumick Chatterjee. (2020, June 19). soumickmj/MRUnder: Initial Release (Version v0.1). DOI: http://doi.org/10.5281/zenodo.3901455.
Figures