4799
Whole-brain high-resolution MRSI at 7T with non-Cartesian FID-ECCENTRIC in glioma patients1University of Geneva, Geneva, Switzerland, 2CIBM Center for Biomedical Imaging, Geneva, Switzerland, 3Medical University of Vienna, Vienna, Austria, 4University of Pittsburgh Medical Center, Pittsburgh, PA, United States, 5Athinoula A. Martinos Center for Biomedical Imaging, Boston, MA, United States, 6Massachusetts General Hospital, Boston, MA, United States, 7Brigham and Women Hospital, Boston, Switzerland
Synopsis
MR spectroscopic imaging (MRSI) can map specific metabolic alterations of glioma brain tumors. Increasing structural details for spatial distribution of metabolites is needed to probe tumor margins and heterogeneity with higher sensitivity and specificity. Here we evaluate the performance of ECCENTRIC, a newly developed non-Cartesian compressed sense MRSI method, to realize fast and high-resolution metabolic imaging at 7T ultra-high field in glioma patients. This is expected to improve diagnosis, prognostication, planning, guidance, and response assessment of treatment in glioma patients and other brain diseases.
Introduction
Magnetic resonance spectroscopic imaging (MRSI) can measure metabolic alterations in glioma for tumor typing and grading but clinical protocols are slow and low resolution1. High-resolution MRSI imaging might improve tumor margins delineation for surgical resection and intra-tumor heterogeneity assessment for treatment response2. Use of standard MRSI techniques3 for high-resolution are prohibitively long and only novel methods of accelerated data acquisition would enable metabolic imaging at high spatial resolution. In this work we assess the performance of ECCENTRIC, a new MRSI method that combines fast non-cartesian random k-space trajectories at 7 Tesla, for fast metabolic imaging of glioma patients. ECCENTRIC (ECcentric Circle ENcoding TRajectorIes for Compressed-sensing) is a technique that aims to dramatically accelerated high-resolution MRSI for clinical use4. ECCENTRIC is a sampling scheme made of circular trajectories randomly positioned in the k-space with several advantages: 1) is not demanding for gradient hardware performance, 2) does not need temporal interleaving, 3) the random distribution of circles enables undersampling of the trajectory without emergence of coherent artifact in the image space. This encoding scheme in combination with low-rank and constrained reconstruction enables the acquisition of metabolic imaging with an acceleration factor of 50-100 compared to usual Cartesian techniques.Methods
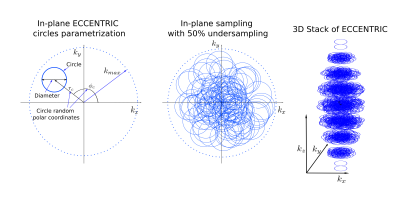
MR acquisition: A short echo free induction decay excitation combined with ECCENTRIC encoding (FID-ECCENTRIC sequence) was implemented on a 7T MRI (Terra, Siemens, Erlangen, Germany) with a NOVA head coil (32Rx/1Tx). The acquisition parameters included: 0.9 ms echo-time, 275 ms repetition-time, 27° excitation flip-angle (SLR pulse, 1ms×6.5kHz), field-of-view (A/P-R/L-H/F) 220×220×105 mm3, 64×63×31 matrix with 3.4 mm isotropic spatial resolution. The ECCENTRIC circle diameter was set to |k_max| / 2 that allowed 2326Hz spectral bandwidth with no temporal interleaving (Figure 1). Acceleration factor 2 by random undersampling resulted in 9min:20s acquisition time. MRSI data were reconstructed with a low-rank compressed-sense model5 and metabolic maps were created after spectral fitting with LCModel6. Water reference signal was acquired with the same parameters but a rosette trajectory without acceleration and a smaller matrix size (22×22×19) in 1min:10s. Anatomical images were acquired with MP2RAGE at 1 mm isotropic resolution and FLAIR at 1.0×1.0×3.0 mm³ resolution for structural anatomy.Glioma patients demographics: 11 glioma patients (3 GBM, 2 anaplastic astrocytoma, 1 anaplastic oligodendroglioma, 2 astrocytoma, 3 oligodendroglioma; 10 mutant IDH; 5 19/19q codeleted; age 28-62; 8M/3F) were scanned with informed consent.
Metabolite content assessment: To evaluate the FID-ECCENTRIC performance to detect tumor metabolic alterations, a region-of-interest (ROI) was delineated using FLAIR on the hyperintense lesion and coregistered to the MRSI volume. A contralateral ROI was similarly created for comparison to the healthy tissue. For each MRSI voxel in the ROIs, only metabolites with Cramer-Rao Lower Bound (CRLB) < 30%, linewidth < 0.1 ppm and SNR >3 were used for the analysis. The differences of metabolite ratio to tCre between the tumor and healthy tissue were compared with a paired t-testtest. Also, the distribution of the quality parameters: CRLB, SNR and linewidth were reported for both tumor and healthy tissue.
Results
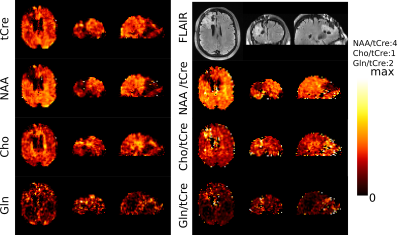
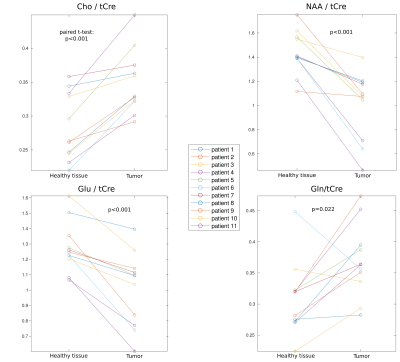
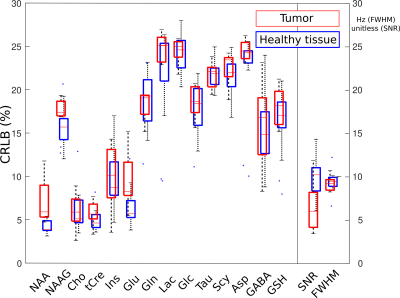
Patient metabolic imaging shown in Figure 2 illustrate the performance of FID-ECCENTRIC. Anatomical and pathological landmarks are visible in the metabolic images at 3.4 mm isotropic resolution. The quantitative comparison in figure 3 showed a significant increase of Cho/tCre and Gln/tCre in tumor while NAA/tCre and Glu/tCre are decreased in agreement with previously published results7. Figure 4 shows the distribution of the quality parameters (CRLB, LW, SNR) for all metabolites which is within acceptable limits for quantitative analysis in majority (95%) of the voxels.Discussion
In this study we investigate the application of FID-ECCENTRIC MRSI for the metabolic imaging of glioma patients. The 3D metabolite mapping in 3.4mm isotropic resolution shows high sensitivity and specificity to pathological tissue, and exhibits significant contrast with healthy tissue.Acknowledgements
No acknowledgement found.References
1. Maudsley, A.A., et al., Advanced magnetic resonance spectroscopic neuroimaging: Experts' consensus recommendations. NMR Biomed, 2020: p. e4309.
2. Bai, J., J. Varghese, and R. Jain, Adult Glioma WHO Classification Update, Genomics, and Imaging: What the Radiologists Need to Know. Top Magn Reson Imaging, 2020. 29(2): p. 71-82.
3. Oz, G., et al., Clinical proton MR spectroscopy in central nervous system disorders. Radiology., 2014. 270(3): p. 658-79.
4. Klauser, A., et al. ECcentric Circle ENcoding TRajectorIes for Compressed-sensing (ECCENTRIC): A fully random non-Cartesian sparse Fourier domain sampling for MRSI at 7 Tesla. ISMRM annual meeting 2021, # 0835 .
5. Klauser, A., et al., Achieving high-resolution (1)H-MRSI of the human brain with compressed-sensing and low-rank reconstruction at 7 Tesla. J Magn Reson, 2021. 331: p. 107048.
6. Provencher, S.W., Estimation of Metabolite Concentrations from Localized in-Vivo Proton Nmr-Spectra. Magnetic Resonance in Medicine, 1993. 30(6): p. 672-679.
7. Hangel, G., Cadrien, C., Lazen, P., Furtner, J., Lipka, A., Hečková, E., … Bogner, W. (2020). High-resolution metabolic imaging of high-grade gliomas using 7T-CRT-FID-MRSI. NeuroImage: Clinical, 28, 102433. https://doi.org/10.1016/j.nicl.2020.102433
Figures