4797
A versatile framework for chemical species separation in Dixon MR Fingerprinting1Department of Diagnostic and Interventional Radiology, School of Medicine, Technical University of Munich, Munich, Germany, 2Philips Research Lab, Hamburg, Germany, 3Philips Healthcare, Hamburg, Germany, 4Department of Diagnostic and Interventional Neuroradiology, School of Medicine, Technical University of Munich, Munich, Germany
Synopsis
Dixon Magnetic Resonance Fingerprinting (Dixon-MRF) is being increasingly used to measure multiple quantitative parameters with effective fat suppression across body tissues. Dixon-MRF processing is typically performed in two steps: water-fat separation and dictionary matching. The present work introduces a versatile formulation for chemical species separation in Dixon-MRF (CSS-MRF) which allows to process a multi-echo fingerprint in a single step obtaining a water fingerprint, a fat fingerprint, a B0 value and a R2* value as outputs. CSS-MRF allows an easy and flexible definition of the parameters to be estimated and enables the option of a varying fat spectrum across the MRF-dimension.
Purpose
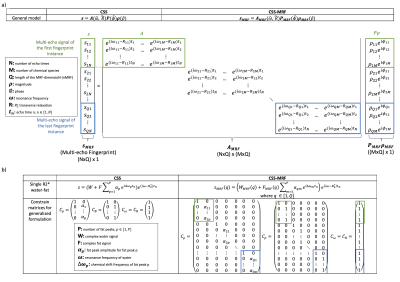
Magnetic resonance fingerprinting (MRF) enables the simultaneous acquisition of several quantitative parameters1 and has been increasingly used in applications outside the brain, including the prostate2, the abdomen3, the myocardium4 and the breast5. However, in body applications, MRF acquisitions require efficient fat suppression. Dixon Magnetic Resonance Fingerprinting (Dixon-MRF) enables water-fat separation to remove the fat-induced blurring in spiral imaging6,7, to remove bias in water relaxometry8,9 and to quantify the tissue fat fraction10. Dixon-MRF processing is typically performed in two steps: water-fat separation and subsequent dictionary matching (Figure 1a). The water-fat separation step relies on $$$B_0$$$ and $$$R_2^*$$$ maps which are unique for every voxel in the image along the MRF-dimension (nMRF). The $$$B_0$$$ map has been previously acquired from a pre-scan6, estimated from averaging the data over nMRF7, included in the dictionary avoiding the Dixon-MRF step9 or estimated together with the $$$R_2^*$$$ map from singular images of all echo times4,10. The pre-scans or estimated $$$B_0$$$ and $$$R_2^*$$$ maps are then used to solve the water fat signals, performing the water-fat separation as many times as the length of the MRF-dimension.This work presents a versatile formulation for chemical species separation11 in Dixon-MRF (CSS-MRF) which allows to process a multi-echo fingerprint of one voxel in a single step with a water fingerprint, a fat fingerprint, a $$$B_0$$$ and a $$$R_2^*$$$ value as outputs (Figure 1b). CSS-MRF allows an easy and flexible definition of the parameters to be estimated; of the fat spectrum, letting the option of a varying fat spectrum across the MRF-dimension available; and aims to set the basis towards the estimation of $$$B_0$$$ and $$$R_2^*$$$ from MRF data.
Methods
As shown in Figure 2a, based on the multi-echo gradient-echo MR signal model $$$s(t_n) = \sum_{m=1}^{M} \rho_m e^{i\phi_m} e^{(i\omega_m-R_m)t_n}$$$, CSS (11) explores its matrix formulation $$$s=A(\omega, R)P(\phi)\rho(\rho)$$$ and derives a flexible way to define the parameters to be estimated using constraint matrices that capture all parameter relations. The measurements at different echo times at a fingerprint instance ($$$MRF_i$$$) represent the signal $$$s$$$ and have their corresponding $$$A, P$$$ and $$$\rho$$$. In CSS-MRF, the $$$s$$$ signals are stack together as a multi-echo fingerprint, $$$s_{MRF}$$$; $$$P\rho$$$ is grouped as a multi-species fingerprint, $$$P_{MRF}\rho_{MRF}$$$; and the $$$A$$$ matrices are organized diagonally to form $$$A_{MRF}$$$. This organization allows to follow the minimization steps of the traditional CSS. Figure 2b presents the formulations for a single $$$R_2^*$$$ water-fat model. $$$C_\rho$$$ indicates that the water has a single peak and the fat, multiple peaks related by the $$$\alpha_p$$$. $$$C_\phi$$$ defines one phase for water and one phase for all fat peaks. $$$C_\omega$$$ and $$$C_{R}$$$ restricts the $$$B_0$$$ and $$$R_2^*$$$ values to be unique for all chemical species.For testing this application, multi-echo gradient-echo cartesian MRF images were acquired from a PDFF phantom containing 15 vials with different fat fractions (Calimetrix, WI, USA) and a volunteer. The multi-echo spoiled gradient-echo sequence with a monopolar readout gradient (12) was used to acquire 16 echoes (TR=40ms, TE1=5ms, dTE=1.3ms) with a MRF flip angle train varying smoothly along nMRF (Figure 1c). The average over nMRF7 was calculated obtaining one image per echo. 6 echoes were then processed by traditional CSS to estimate the $$$B_0$$$ and $$$R_2^*$$$ maps. Finally, 6 echoes of the acquired signal were processed by CSS-MRF. For the phantom, the fat spectrum was estimated from a MRS scan. The RenSCAT13 fat model was applied in-vivo.
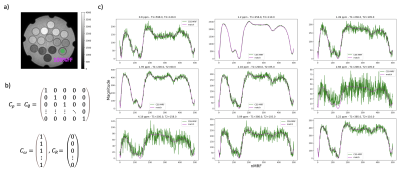
A second application of this framework is the fat spectrum characterization along nMRF. Dixon-MRF considers a constant fat model along nMRF, which could be not the case specially at high FF12. The fat spectrum of the 100% FF phantom’s vial was estimated using CSS-MRF and based on the respective constraint matrices.
Results
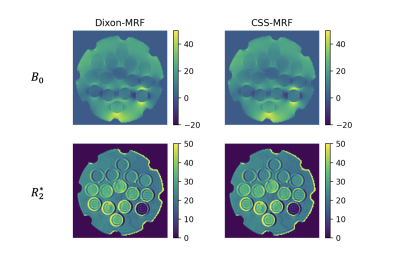
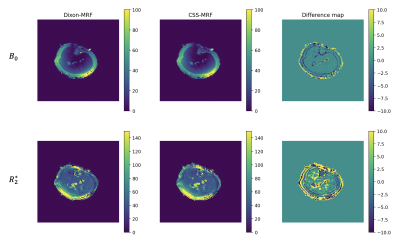
CSS-MRF reports equivalent results to Dixon-MRF in phantom and in vivo images. Figure 3 presents a comparison of the $$$B_0$$$ and $$$R_2^*$$$ maps in the phantom estimated from averaging the data over nMRF and using CSS-MRF. The agreement of the maps confirms an equivalent performance of the CSS-MRF to the previously proposed Dixon-MRF processing. Figure 4 similarly presents an equivalent performance in vivo leg data.Figure 5 shows the decomposition of a fat fingerprint into its fat peak fingerprints using CSS-MRF. With this, the fat spectrum along nMRF can be calculated and used further for water fat separation varying the fat model per $$$MRF_i$$$, but still with the restriction of a common $$$B_0$$$ and $$$R_2^*$$$ values. This complex scenario is easy to formulate using CSS-MRF by defining the different fat spectrum models in each part of $$$C_\rho$$$, similar to Figure 2b.
Discussion and conclusion
CSS-MRF has shown its versatility for constraining parameters in the water-fat separation step of Dixon-MRF. The benefit of obtaining $$$B_0$$$ and $$$R_2^*$$$ maps in a single step differences CSS-MRF from the traditional approaches. Furthermore, its flexibility also helps with the fat spectrum characterization along nMRF and its further application because CSS-MRF provides a framework where every $$$MRF_i$$$ can have a different fat spectrum, but all share a unique $$$B_0$$$ and $$$R_2^*$$$ value. This should be reviewed for further investigation on the effect of the fat spectrum in MRF, especially for undersampled MRF acquisitions.Acknowledgements
The present research was supported by Philips Healthcare.References
- Ma D, Gulani V, Seiberlich N, Liu K, Sunshine JL, Duerk JL, Griswold MA.Magnetic resonance fingerprinting. Nature 2013;495(7440):187-192.
- Yu AC, Badve C, Ponsky LE, Pahwa S, Dastmalchian S, Rogers M, Jiang Y, Margevicius S, Schluchter M, Tabayoyong W, Abouassaly R, McGivney D, Griswold MA, Gulani V. Development of a Combined MR Fingerprinting and Diffusion Examination for Prostate Cancer. Radiology 2017;283(3):729-738.
- Chen Y, Jiang Y, Pahwa S, Ma D, Lu L, Twieg MD, Wright KL, Seiberlich N, Griswold MA, Gulani V. MR Fingerprinting for Rapid Quantitative Abdominal Imaging. Radiology 2016;279(1):278-286.
- Jaubert O, Cruz G, Bustin A, Schneider T, Lavin B, Koken P, Hajhosseiny R, Doneva M, Rueckert D, Botnar RM, Prieto C. Water-fat Dixon cardiac magnetic resonance fingerprinting. Magn Reson Med 2020;83(6):2107-2123.
- Panda A, Chen Y, Ropella-Panagis K, Ghodasara S, Stopchinski M, Seyfried N, Wright K, Seiberlich N, Griswold M, Gulani V. Repeatability and reproducibility of 3D MR fingerprinting relaxometry measurements in normal breast tissue. J Magn Reson Imaging 2019;50(4):1133-1143.
- Koolstra K, Webb AG, Veeger TTJ, Kan HE, Koken P, Bornert P. Water-fat separation in spiral magnetic resonance fingerprinting for high temporal resolution tissue relaxation time quantification in muscle. Magn Reson Med 2020;84(2):646-662.
- Nolte T, Gross-Weege N, Doneva M, Koken P, Elevelt A, Truhn D, Kuhl C, Schulz V. Spiral blurring correction with water-fat separation for magnetic resonance fingerprinting in the breast. Magn Reson Med 2020;83(4):1192-1207.
- Ostenson J, Damon BM, Welch EB. MR fingerprinting with simultaneous T1, T2, and fat signal fraction estimation with integrated B0 correction reduces bias in water T1 and T2 estimates. Magn Reson Imaging 2019;60:7-19.
- Marty B, Carlier PG. MR fingerprinting for water T1 and fat fraction quantification in fat infiltrated skeletal muscles. Magn Reson Med 2020;83(2):621-634.
- Jaubert O, Arrieta C, Cruz G, Bustin A, Schneider T, Georgiopoulos G, Masci PG, Sing-Long C, Botnar RM, Prieto C. Multi-parametric liver tissue characterization using MR fingerprinting: Simultaneous T1 , T2 , T2 *, and fat fraction mapping. Magn Reson Med 2020;84(5):2625-2635.
- Diefenbach MN, Liu C, Karampinos DC. Generalized parameter estimation in multi-echo gradient-echo-based chemical species separation. Quant Imaging Med Surg 2020;10(3):554-567.
- Huaroc E, Weidlich D, Amthor T, Koken P, Baumann M, Weiss K, Makowski M, Schwaiger J, Doneva M, Karampinos D. On the effect of fat spectrum complexity in Dixon MR Fingerprinting. Proc. Intl. Soc. Mag. Reson. Med. 27 (2021) Abstract 5544
- Ren J, Dimitrov I, Sherry AD, Malloy CR. Composition of adipose tissue and marrow fat in humans by 1H NMR at 7 Tesla. J Lipid Res 2008;49(9):2055-2062
Figures
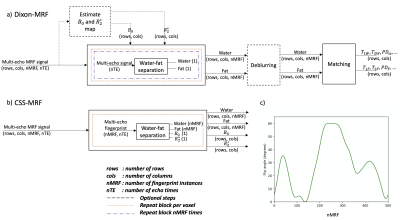
Figure 1. a) Traditional Dixon-MRF approach. The water-fat separation is repeated per voxel and per fingerprint instance. B0 and R2* maps are obtained from a pre-scan or estimated from averages over the MRF dimension. The application of the deblurring step depends on the type of MRF data. Therefore, both steps are represented as optional. b) CSS-MRF performs the water-fat separation of a fingerprint in a single step including B0 and R2* as outputs. c) Flip angle train used for the experiments.