4795
Deep learning Circle of Willis arterial labeling strategies for pipeline of cerebrovascular disease analysis1Laboratory of Imaging Technologies, Faculty of Electrical Engineering, University of Ljubljana, Ljubljana, Slovenia, 2Division of Interventional Neuroradiology, Department of Radiological Sciences, Ronald Reagan UCLA Medical Center, David Geffen School of Medicine at UCLA, Los Angeles, CA, United States
Synopsis
The correlation between different variants of the Circle of Willis (CoW) and cerebrovascular disorders such as stroke, aneurysms and mental disorders is not yet well understood. A step towards a better understanding of the role of CoW in the aforementioned diseases is the automatic labeling of main vessels. In this work we tested three different approaches and observed high mIoU value of 0.870. As such the automatic anatomical labelling of the CoW seems feasible for clinical evaluation of the association of different anatomical variants with the risk factors of cerebrovascular pathologies.
Introduction
The Circle of Willis (CoW) is the bridging of the left and right anterior cerebral circulation with the posterior circulation, with the role of ensuring unrestricted blood flow throughout the whole brain1. Variations of the CoW are common, including hypoplastic, missing, or duplicated arteries, and it is estimated that 52% of people lack at least one segment2,3. Although the role of different variants are not yet well understood, they can play an important role in the occurrence, manifestation of symptoms, treatment options and recovery process of cerebrovascular disorders such as stroke, aneurysms and mental disorders2,4,5.Arterial labeling of the CoW is a prerequisite for geometric characterization of the CoW, and in practice is often performed manually. This is a monotonous and time-consuming job, which relies on radiological imaging experts. The purpose of this study was to automate and validate anatomical labeling of the CoW, using recent deep learning approaches. Quick automatic labeling would allow accurate and consistent geometric characterization of the CoW in large-scale studies and therefore enable the analysis of anatomy based risk factors associated with cerebrovascular pathologies.Methods
This study focused on 3D time-of-flight magnetic resonance angiography (TOF MRA) scans, a standard clinical method for neurovascular imaging. Images of healthy subjects were obtained from the publicly available IXI dataset6. The labeling dataset was generated by first segmenting the CoW and surrounding vessels using interactive thresholding, followed by visual check. If required, the segmentation of tiny communicating arteries were manually corrected. The resulting manual segmentations were transformed into trigonometric meshes7, and individual mesh vertices were labeled by CoW artery (ICA, ACA, PCA, BA, PCo or SCA). Vascular mesh vertex data was used to train three state-of-the-art deep learning models; namely, PointNet (PN)8, PointNet++ with multiscale grouping (PN++ MSG)9 and Dynamic Graph Convolutional Neural Network (DGCNN)10. The three models were applied as part segmentation methods to assign one of the CoW labels to each mesh vertex. The obtained vertex labels were compared to the manually assigned labels using the mean intersection over union (mIoU) metric, with values closer to one indicating more accurate anatomical labeling of the CoW. 81 manually segmented MRA scans were used to test the models. The mIoU was computed using cross-validation with 80/20 train/test dataset split.Results
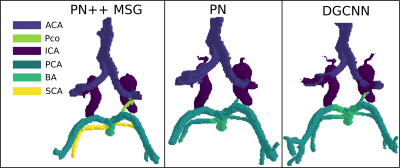
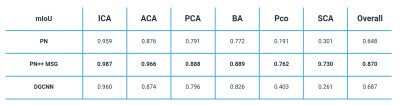
Table 1 shows the mIoU per each CoW segment, and overall, for the three tested methods, while an example of labelled CoW of the same subject obtained from the three tested methods is shown in Figure 1. The PN++ model achieved the best results, followed by the PN and DGCNN. The PN and DGCNN methods were clearly inferior, which was most evident in the labeling of the smaller PCo and SCA. The PN and DGCNN methods often did not identify the SCA segment, which is very similar in position and shape to the PCA artery (see Figure 1). As a result, the majority of falsely labelled vertices were false positive PCA labels. Furthermore, the mIoU values of individual CoW segments were proportional to their size and specific shape. For instance, even using the PN++ MSG, the PCo segment had a relatively low mIoU score of 0.762, due to smaller vessel radii and shorter length (on average), and thus fewer mesh vertices, as compared to other CoW segments. The overall high accuracy of PN++ MSG (mIoU=0.870), compared to the other two methods, was due to its hierarchical multiscale information aggregation taking into account the structure of surrounding veins that indirectly contribute to CoW part recognition.Discussion
This study shows that deep learning part segmentation method based on PN++ MSG can be successfully applied to automatic labeling of the CoW. While state-of-the-art methods are based on heuristic vessel topology analysis, they exhibit clear disadvantages such as the need for manual interaction11,12 to mark certain artery segments and thus obtain a vessel graph. Robben et al.13 proposed an automated approach, however, it requires substantial compute and time resources to extract the vessel topology and thus seems less suitable for large scale studies.Among the tested methods, the PN++ MSG achieved the best mIoU of 0.870 mIoU, and visually correct anatomical labeling in all cases. For comparison, Bogunovič et al.11 achieved correct vessel labelling in 58% of cases. Due to different evaluation procedures the results can not be directly compared to ours, as in the mentioned works the authors evaluated the labelling accuracy on centerlines and and bifurcations, not on the actual topology of cerebral vessels. Furthermore, the inconsistencies of the manual labelling, especially in localizing the appropriate distal vessel cut-offs may affect the validation of anatomical labelling. In general, the 3D deep learning method PN++ MSG proved to be very accurate and robust and executed in about 10 seconds, on average; hence, its seems feasible to apply in large scale studies to obtain accurate and consistent anatomical labelling.Conclusion
Among the three tested automatic methods the PN++ MSG was capable of accurate vessel segment labelling of the CoW, based on the observed with high mIoU value of 0.870. As such the automatic anatomical labelling of the CoW seems feasible for clinical evaluation of the association of different anatomical variants with the risk factors of cerebrovascular pathologies.Acknowledgements
This study was supported by the Slovenian Research Agency (Core Research Grant No. P2-0232 and Research Grants Nos. J2-8173 and J2-2500).
This research is inpart supported by NIH R01HL152270, AHA 18IPA34170130 and UCLA Exploratory Research Grant.