4787
Simultaneous learning of group-wise registration and joint segmentation of cardiac cine MRI with sparse annotation1Department of Computer Science and Technology, University of Cambridge, Cambridge, United Kingdom, 2School of Biomedical Engineering, ShanghaiTech University, Shanghai, China, 3Department of Interventional and Diagnostic Radiology, University Hospital of Tuebingen, Tuebingen, Germany, 4Shanghai United Imaging Intelligence Co., Ltd., Shanghai, China
Synopsis
Numerous deep learning methods have been proposed for cardiac cine MRI segmentation, while most of them require laborious annotation for supervised training. Herein, we propose an approach to perform group-wise registration and joint segmentation of cardiac cine images, by training a registration network in a self-supervised manner to align dynamic images to their mean image space and also a segmentation network in a weakly-supervised manner using sparsely-annotated data and predicted motions from the registration network. By training these two (registration and segmentation) networks simultaneously, our proposed joint learning approach provides better segmentations than the direct segmentation network.
Introduction
Cardiac motion estimation and segmentation on cardiac cine MRI are important tasks for cardiac function analysis1-3. Deep learning-based cardiac segmentation has been extensively investigated, while most of the proposed methods require supervised training4-10. Recently, self-supervised deep learning-based cardiac motion estimation is shown feasible by enforcing warped image similarity and motion smoothness11, and has been employed to aid the segmentation task7. However, current deep learning methods adopt pair-wise registration which may have poor registration performance between diastolic and systolic frames when cardiac motion is large. To address the above challenges, we propose an approach that jointly learns group-wise registration and segmentation, where the registration network is trained in a self-supervised manner to register the dynamic images to their mean space avoiding registering between end-diastolic (ED) and end-systolic (ES) frames, and the segmentation network is trained weakly-supervised with sparsely annotated data (only ED and ES frames are labeled) by taking advantage of the predicted motion. The two networks are trained simultaneously with a composite loss. The effectiveness of the joint learning approach is validated against the network that considers segmentation only.Methods
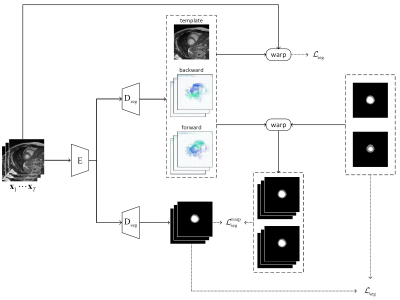
Network ArchitectureAs shown in Fig. 1, the joint learning framework consists of two branches: 1) the self-supervised registration branch that registers the dynamic images to an implicit template which is the mean space of the dynamic sequence, and 2) the semi-supervised segmentation branch that segments the left ventricle (LV) and myocardium (Myo). Both branches use the adapted U-Net architecture12 and share a common encoder.
In the registration branch, given a sequence of cardiac MR images $$$\mathbf{x}_1,...,\mathbf{x}_T$$$, the group-wise registration network aims to estimate the transformations that warp the image sequence to an implicit template $$$\mathbf{x}_\text{temp}$$$. The transformations are constrained to be diffeomorphic by estimating the velocity fields $$$\mathbf{v}_1,...,\mathbf{v}_T$$$ from the template to the dynamic images. The forward and backward motion fields can be estimated by integrating the velocity field over unit time13. The spatial transformer14 is applied to encode the motion and warp an image from the source to the target frame using linear interpolation.
To optimize the network, mean-squared error loss is used between: 1) the implicit template $$$\mathbf{x}_\text{temp}$$$ and the warped template from the original sequence, and 2) the warped sequence from the implicit template $$$\{\mathbf{\hat{x}}^{(1)}_\text{temp},...,\mathbf{\hat{x}}^{(T)}_\text{temp}\}$$$ and the original image sequence. In addition, a diffusion regularization on velocity fields is included to encourage smoothness. The registration loss can be formulated as: $$\mathcal{L}_\text{reg}=\frac{1}{T}\sum_{i=1}^T ||\mathbf{x}_\text{temp}-\mathbf{\hat{x}}^{(i)}_\text{temp}||^2+||\mathbf{\hat{x}}_i-\mathbf{x}_i||^2+\lambda \text{TV}(\mathbf{\nabla v}),$$ where a regularization parameter $$$\lambda$$$ of 0.1 is used.
In the segmentation branch, a categorical cross-entropy loss is firstly applied to train the network on labeled data as $$$\mathcal{L}_\text{seg}=-\sum_{i \in L} \mathbf{y}_i\text{log}(f_\theta(\mathbf{x}_i))$$$, where $$$L$$$ is the indices of annotated image frames, $$$\mathbf{y}_i$$$ is the ground-truth segmentation masks, and $$$f_\theta$$$ is the segmentation network parameterized by $$$\theta$$$. Furthermore, additional supervision is introduced from the predicted motion by warping the ground-truth segmentation masks to generate the masks for other unannotated frames. Then, another categorical cross-entropy loss can be similarly defined as $$$\mathcal{L}_\text{seg}^\text{warp}=-\sum_{i\in U}\mathbf{y}_i^{w}\text{log}(f_\theta(\mathbf{x}_i))$$$, where $$$U$$$ is the indices of unannotated image frames, and $$$\mathbf{y}_i^{w}$$$ is the warped segmentation masks.
Training and Validation
We evaluate our method on the Automatic Cardiac Diagnosis Challenge (ACDC) dataset, consisting of cine-MR images and the corresponding segmentation masks of ED and ES frames of 100 patients. The preprocessed dataset with 280 slices is randomly split with a ratio of 5:1:1 for training, validation, and testing. All networks are optimized using the Adam optimizer with the learning rate of $$$10^{-4}$$$. The registration network is firstly trained for 100 epochs, and then both networks are trained for another 100 epochs.The joint learning approach is compared with training only the segmentation branch (seg-only) to evaluate its effectiveness. We manually segment all the dynamic frames in the testing data as the ground truth, and then the Dice coefficient and Hausdorff distance are calculated between the predicted segmentation and the ground truth to evaluate the performance.
Results
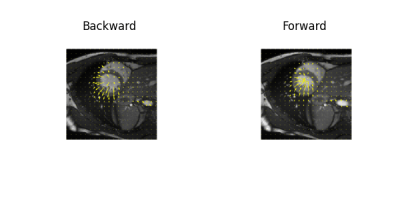
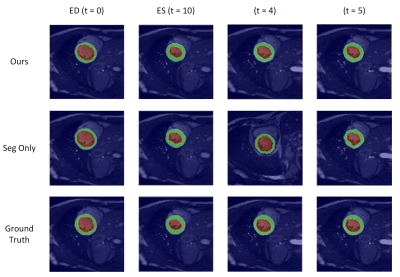
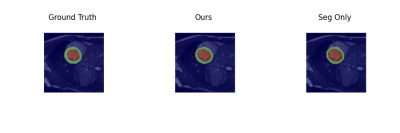
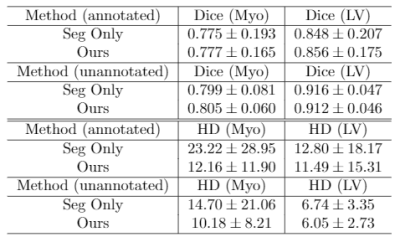
Examples of estimated motion and the learned template image are shown in Fig. 2. It can be seen the group-wise registration network learns smooth and plausible cardiac motion. Typical segmentation results for ED, ES and two other unlabeled frames with the proposed joint learning approach and seg-only approach with our manual segmentations are shown in Fig. 3. The joint learning approach provides segmentations closer to the manual annotations, especially for non-ED and non-ES frames. The segmentation results for a whole dynamic sequence are provided in Fig. 4. The joint learning approach generates visually better segmentations. Table. 1 summarizes the segmentation performance of our method and the baseline in the test set. It can be seen that the joint learning approach achieves slightly higher Dice coefficient, but its Hausdorff distance is much lower than the seg-only method.Conclusion
Our proposed joint learning of group-wise registration and segmentation framework can be efficiently optimized to produce high-quality segmentation masks on partially annotated datasets. The introduction of a group-wise registration branch trained with self-supervision to optimize simultaneously the motion field and the reference template provides additional supervision to the segmentation branch. The group-wise registration can also be combined with other more advanced segmentation networks to improve the segmentation performance.Acknowledgements
No acknowledgement found.References
[1] M. Salerno, B. Sharif, H. Arheden, A. Kumar, L. Axel, D. Li, and S. Neubauer, “Recent advances in cardiovascular magnetic resonance: techniques and applications,” Circulation: CardiovascularImaging, vol. 10, no. 6, p. e003951, 2017.
[2] F. H. Epstein, “Mri of left ventricular function,” Journal of nuclear cardiology, vol. 14, no. 5, pp. 729–744, 2007.
[3] C. M. Kramer, J. Barkhausen, S. D. Flamm, R. J. Kim, and E. Nagel, “Standardized cardiovascular magnetic resonance (cmr) protocols 2013 update,” Journal of Cardiovascular Magnetic Resonance, vol. 15, no. 1, pp. 1–10, 2013.
[4] J. Duan, G. Bello, J. Schlemper, W. Bai, T. J. Dawes, C. Biffi, A. de Marvao, G. Doumoud, D. P.O’Regan, and D. Rueckert, “Automatic 3d bi-ventricular segmentation of cardiac images by a shape-refined multi-task deep learning approach,” IEEE transactions on medical imaging, vol. 38, no. 9, pp. 2151–2164, 2019.
[5] O. Oktay, E. Ferrante, K. Kamnitsas, M. Heinrich, W. Bai, J. Caballero, S. A. Cook, A. De Marvao, T. Dawes, D. P. O‘Regan, et al., “Anatomically constrained neural networks (acnns): application to cardiac image enhancement and segmentation,” IEEE transactions on medical imaging, vol. 37, no. 2, pp. 384–395, 2017.
[6] C. Zotti, Z. Luo, A. Lalande, and P.-M. Jodoin, “Convolutional neural network with shape prior applied to cardiac mri segmentation,” IEEE journal of biomedical and health informatics, vol. 23, no. 3, pp. 1119–1128, 2018.
[7] C. Qin, W. Bai, J. Schlemper, S. E. Petersen, S. K. Piechnik, S. Neubauer, and D. Rueck-ert, “Joint learning of motion estimation and segmentation for cardiac mr image sequences,” in International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 472–480, Springer, 2018.
[8] C. Qin, W. Bai, J. Schlemper, S. E. Petersen, S. K. Piechnik, S. Neubauer, and D. Rueckert, “Jointmotion estimation and segmentation from undersampled cardiac mr image,” inInternationalWorkshop on Machine Learning for Medical Image Reconstruction, pp. 55–63, Springer, 2018.
[9] J. Liu, H. Liu, S. Gong, Z. Tang, Y. Xie, H. Yin, and J. P. Niyoyita, “Automated cardiac segmentation of cross-modal medical images using unsupervised multi-domain adaptation and spatial neural attention structure,” Medical Image Analysis, p. 102135, 2021.
[10] N. Painchaud, Y. Skandarani, T. Judge, O. Bernard, A. Lalande, and P.-M. Jodoin, “Cardiacsegmentation with strong anatomical guarantees,” IEEE Transactions on Medical Imaging, vol. 39, no. 11, pp. 3703–3713, 2020.
[11] M. A. Morales, D. Izquierdo-Garcia, I. Aganj, J. Kalpathy-Cramer, B. R. Rosen, and C. Catana, “Implementation and validation of a three-dimensional cardiac motion estimation network,” Radiology: Artificial Intelligence, vol. 1, no. 4, p. e180080, 2019.
[12] O. Ronneberger, P. Fischer, and T. Brox, “U-net: Convolutional networks for biomedical image segmentation,” in International Conference on Medical image computing and computer-assisted intervention, pp. 234–241, Springer, 2015.
[13] A. V. Dalca, G. Balakrishnan, J. Guttag, and M. R. Sabuncu, “Unsupervised learning of probabilistic diffeomorphic registration for images and surfaces,” Medical image analysis, vol. 57, pp. 226–236, 2019.
[14] M. Jaderberg, K. Simonyan, A. Zisserman,et al., “Spatial transformer networks,” Advances in neural information processing systems, vol. 28, pp. 2017–2025, 2015.
Figures