4785
Automated Time Adjusted Inversion Time Selection for Late Gadolinium Enhancement Imaging - Validation and Application on Patient Datasets1Pattern Recognition Lab, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany, 2Magnetic Resonance, Siemens Healthcare GmbH, Erlangen, Germany, 3Siemens Medical Solutions USA, Inc, Princeton, NJ, United States, 4Department of Radiology, University Medical Center, Johannes Gutenberg-University Mainz, Mainz, Germany, 5Division of Cardiovascular Imaging, Department of Radiology and Radiological Science, Medical University of South Carolina, Charleston, Germany, 6Diagnostikum Berlin, Berlin, Germany, 7Department of Cardiology, Robert Bosch Medical Center, Stuttgart, Germany, 8Siemens Healthcare France, Saint-Denis, France, 9Institut Cardiovasculaire Paris Sud, Cardiovascular Magnetic Resonance Laboratory, Hôpital Privé Jacques Cartier - Ramsay Santé, Massy, France, 10Division of Cardiology, Johns Hopkins University, Baltimore, MD, United States
Synopsis
In cardiac MRI, the Late Gadolinium Enhancement technique is usually performed after inversion recovery scout sequences that are acquired to null the myocardial properly for optimal image contrast. In clinical practice, the selection and adjustment of the inversion time for healthy myocardium nulling are manually performed. To standardize and automate the process, we propose an automated deep-learning-based system combined with a linear regression model, which automates the selection of the inversion time, taking into consideration the time delay between the inversion time scout and LGE sequences. In this work, we validated the system in a large retrospective study (N=765).
Introduction
In cardiac MRI, late gadolinium enhancement (LGE), a contrast enhanced inversion recovery sequence is generally performed to assess myocardial viability or to investigate the etiology of cardiomyopathies1-3. In current clinical practice, inversion time (TI) selection for correct myocardial nulling is performed by visual inspection of a TI scout sequence, or a manual post-processing step in case of PSIR LGE. As the contrast agent (CA) concentration within the myocardium changes over time, the TI value determined from a scout sequence at one time point may need to be adjusted for LGE acquisitions at later time points, with literature suggesting differences of up to 50 ms4. We proposed a fully automated system consisting of a deep-learning-based system5-7 that outputs the time with darkest myocardium (TInull) and linear regression model that adjusts the TInull to output TIadjusted by considering the duration between the TI scout series and LGE imaging. In this work, we validated the system in a large retrospective study (N=765).Methods
Datasets from three different institutions (Institution A to C) acquired on a 1.5T (MAGNETOM Aera, Siemens Healthcare, Erlangen, Germany) and three 3T scanners (MAGNETOM Prisma, Skyra, and Vida systems, Siemens Healthcare, Erlangen, Germany) were used to perform the linear regression analysis. In each of these training datasets, the user-selected actual TI (TIstatic), where the users have made the time adjustment through expert knowledge for LGE imaging, is compared with the TInull for the analysis. An independent set of data (Institution D), gathering a total of 765 datasets, acquired on the 1.5T scanner (MAGNETOM Aera, Siemens Healthcare, Erlangen, Germany) were used to evaluate the performance of the proposed system, with four LGE views (short-axis, 2-3-4 chamber views) performed at different times after CA administration. All TI scout images were acquired with a segmented inversion recovery CINE TrueFISP pulse sequence either with, or without compressed sensing. Detailed information about the datasets is presented in Fig. 1.The proposed prototype system is built with deep-learning-based models5-7 and a linear regression model (Fig. 2). The input to the system is a TI scout sequence of a mid-ventricular short-axis (SAX) slice. Based on the deep-learning models, the TInull is selected from the mean pixel intensities of the myocardium at each time point. Under the assumption of a linear adjustment of TInull for the TIstatic to account for the time gap between the TI scout and LGE sequences, a heuristic linear regression model can be modeled as:
$$TI_{adjusted}(t) = a \cdot t + b + TI_{null},$$
where a and b are the parameters of the linear regression model, t is the duration between the TI scout and LGE imaging acquisition time in minutes, and TInull is the system selected TI with darkest myocardium at the time of the TI scout acquisition.
To validate the system, mean and standard deviation of the absolute difference between each TIstatic and TIadjusted were analyzed based on Bland-Altmann analysis. Some example cases including outliers were qualitatively validated.
Results
The curve fit with linear regression is plotted in Fig. 3. Based on the regression analysis, the slope is set to 3.2, the offset to 30.1 ms. The mean difference in the large retrospective patient study (N=765) was 0.1 ± 26.8 ms (Fig. 4). In 97.8% of the dataset, the absolute difference between TIstatic and TIadjusted was less than 50 ms. In one study, the absolute difference was more than 100 ms. The importance of TIadjusted compared to TInull can be shown in the first two cases in Fig. 5, where TIadjusted differs from TInull with more than 30 ms, while showing good agreement with TIstatic. In some cases, the TIadjusted was selected as the TI, with better contrast than the one chosen for TIstatic as shown in last two cases in Fig. 5.Discussion
TIadjusted selected using the linear regression model shows high accuracy compared to TIstatic in a large retrospective study. Analysis of the regression model with more data from different institutions would produce more robust results. The TIadjusted shows a clear advantage over TInull with the time gap taken into account, as shown in the first example in Fig. 5, where the time gap is large between the TI scout and LGE acquisitions (>6 min). The outlier cases are often due to the TIstatic being selected too late, with more than 50ms (N=34). In these cases, even if the time gap (~2.7 min) was not large, strong offsets were probably chosen, that lead to slight contrast reduction in the myocardium as shown in Fig. 5. A retrospective study comparing the image quality of LGE images acquired with time adjusted TIadjusted vs. non-adjusted TInull vs. manually selected TIstatic by an expert is desirable.Conclusion
The results demonstrate the feasibility of the proposed system to automatically select the correct TI including a temporal adjustment to account for the delay between TI scout and LGE. It shows great potential for improving standardization of LGE imaging.Acknowledgements
No acknowledgement found.References
1. Kellman, P., and Arai A. E. "Cardiac imaging techniques for physicians: late enhancement." JMRI 2012.
2. Kellman, P., et al. "Phase‐sensitive inversion recovery for detecting myocardial infarction using gadolinium‐delayed hyperenhancement." MRM 2002.
3. Munoz, C., et al. "Motion-corrected 3D whole-heart water-fat high-resolution late gadolinium enhancement cardiovascular magnetic resonance imaging." JCMR 2020.
4. Kramer, C. M., et al. "Standardized cardiovascular magnetic resonance imaging (CMR) protocols: 2020 update." JCMR 2020.
5. Yoon, S. et al.: “A Deep-Learning-based Automated Myocardial Inversion Time Selection for Late Gadolinium Enhancement Imaging”, SCMR 2021.
6. Yoon, S. et al.: “Validation of a Deep-Learning-based Automated Myocardial Inversion Time Selection for Late Gadolinium Enhancement Imaging in a Prospective Study.”, ISMRM 2021.
7. Chitiboi, T. et al.: „Automated ventricular volume and strain quantification with deep learning.“, SCMR 2020.
Figures
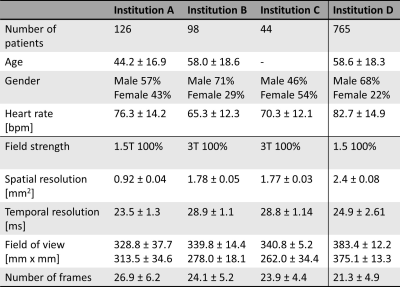
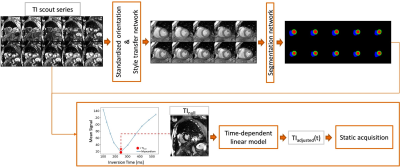
Figure 2: Overview of the proposed system. A SAX TI scout series is used as input to the system. By applying the localization, style transfer and segmentation network, TInull can be selected based on the mean signal intensity of the myocardium. TIadjusted is calculated for the time-dependent linear model for subsequent LGE imaging.
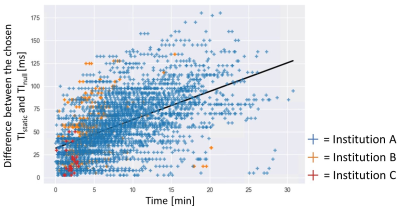
Figure 3: The plot of difference between the chosen TIstatic and TInull over the time gap between the TI scout and LGE acquisitions for the regression analysis. The fit linear model is shown in blue line.
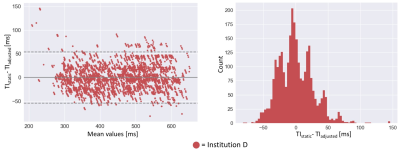
Figure 4: Left: The performance results from the unseen retrospective data between each TIstatic that are acquired after TI scout and the proposed TIadjusted. Right: the univariate histogram plot showing the distribution of the system performance.
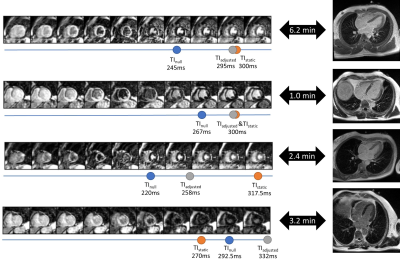
Figure 5: Example cases from the Institution D showing the TI scout series and the resulting LGE images. The automatically selected TInull, TIadjusted and the actual TIstatic used for the subsequent LGE imaging are marked. The time gap between the TI scout and LGE imaging is shown as well. In the first two cases, the TIadjusted deviates from TInull but matched well with TIstatic. The last two cases show the outliers of TIadjusted.