4775
A NOVEL OPEN-SOURCE CARDIAC SEGMENTATION FRAMEWORK FOR CINE MRI1School of Biomedical Engineering, Universidad de Valparaíso, Valparaíso, Chile, 2Department of Radiology, School of Medicine, Pontificia Universidad Católica de Chile, Santiago, Chile, 3Millennium Nucleus in Cardiovascular Magnetic Resonance, CardioMR, Santiago, Chile, 4Institute for Biological and Medical Engineering, Schools of Engineering, Medicine and Biological Sciences, Pontificia Universidad Católica de Chile, Santiago, Chile, 5Biomedical Imaging Center, Pontificia Universidad Católica de Chile, Santiago, Chile
Synopsis
Cardiac contour delineation currently presents several proposals for automatic segmentation in short-axis MRI. However, most of them have drawbacks such as low accuracy with images from other MRI scanners, lack of description of the algorithms, and difficult access for clinical or research use. For this reason, we present VENTSEG, a deep learning framework for cardiac segmentation implemented in Matlab and Python. Our dice coefficient results segmenting images from the CNN validation set are 85.55% and 96.17% on anonymized clinical data. These results demonstrate the generalization of the framework on multidomain images.
Introduction
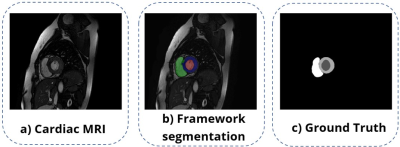
Currently, convolutional neural networks (CNNs) reduce manual interaction and improve the accuracy of cardiac segmentation1-3. However, these machine learning techniques are complex and have high ranges of variability4, causing their prediction efficiency to decline in data from different MRI scanners, acquisition centers, and pathologies5. In this work, we propose to evaluate the performance of a new open-source framework, that we call VENTSEG, based on a deep learning model and several processing algorithms. The framework will allow segmentation of the left ventricle (LV), right ventricle (RV), and myocardium (Myo) contours to be finally compared with their respective ground truth, see Fig. 1.Methods
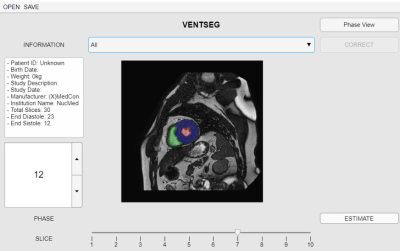
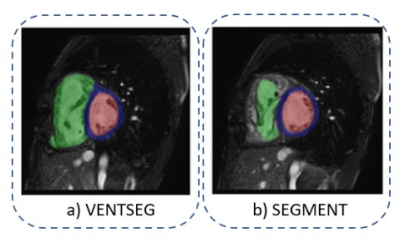
Our first approach tackles the segmentation problem using a CNN. The model used is called Iterative Backpropagation6, which bases its architecture on a U-network, review Ronneberger et al.7. Models were trained for 100 epochs and a batch size of 32 images. The cross-validation method was necessary for the selection of the best model and hyperparameters. The optimizer used was Adam's optimizer with a learning rate of 0.001 and Dice Score for loss calculation. In addition, the model uses two techniques, data augmentation that improves model performance and backpropagation that captures the differences between domains. The data set used in the model was extracted from Campello et al.8, consisting of 300 short-axis images of patients with various cardiomyopathies, four different scanner vendors (GE, Philips, SIEMENS, and Canon), and six clinical institutions. We use 150 datasets for training, 130 for testing, and 20 for validation. Subsequently, we include two stages in the segmentation framework: i) Preprocessing, a stage before CNN consisting of rescaling algorithms, reduction of the plane of interest, normalization and intensity enhancement at ventricular borders; ii) Postprocessing, a stage after the trained model consisting of morphological operations and connected component analysis for error correction. We implemented all these algorithms in a framework with Python script language and MATLAB App Designer graphical interface (Fig. 2). Finally, we use the Dice Coefficient (DC) metric to evaluate the prediction performance of the framework concerning its ground truth. For this purpose, we use the same 20 validation data (9 Philips, 6 GE, 3 Siemens, and 2 Canon). Furthermore, we performed the evaluation of 10 anonymized clinical case data. These data were acquired on a Philips 1.5 T MRI scanner and segmented with the free Segment software (from Medviso)9 to compare their corresponding results (Fig. 3).Results
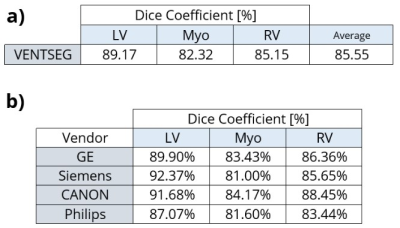
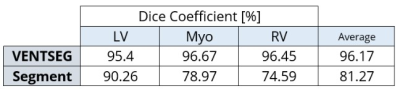
The average DC of the deep learning model according to the validation dataset was 81%. Once we implemented the processing steps, the framework with the same dataset increased its DC to 85.55%. In Figure 4a, we show the segmentation results of each cardiac contours: LV, RV, and Mio, where LV with 89.17% is the cardiac contour with the best prediction performance. Furthermore, we evidenced the segmentation performance of each magnetic resonator brand (Fig. 4b). The performance results of the VENTSEG framework and Segment software based on the clinical dataset are shown in Figure 5. The average DC results are 96.17% and 81.27%, respectively. In this case, our framework presents the highest DC in the Myo contour with 96.67%, while the Segment software in the RV contour presents the lowest DC with 74.59%.Discussion
The DC is the most widely used metric to quantify segmentation performance because it measures the similarity between two images. In this work, we performed a comparison between results obtained by the framework and its ground truth. The results and code are available at Github10. The framework, with processing steps help, achieved to increase its DC by 4.55%. A limitation of our study is realignment and not having a correct reference of the apex or base of the heart because we did not use long-axis or four-chamber views. The segmentation performance for each brand showed that the framework obtains a similarity higher than 80% in all cases and cardiac contours. On the other hand, the performance test with a new database allowed us to verify if the software can correctly segment untrained or untested data. This evidence shows the importance of training the CNN with a robust database. In addition, the last comparison was limited to Segment because it presents less variability than other commercial software available11.Conclusion
Evaluation of our cardiac segmentation framework, using the CD metric, provides consistent and generalizable results. Our results had excellent segmentation predictions on multidomain image sets. Importantly, for future work, we are looking for our segmentation framework to demonstrate reduced user interaction times due to improved accuracy and automation of all user processes.Acknowledgements
ANID - Millennium Science Initiative Program - NCN17_129, ANID FONDECYT de iniciación en investigación FONDECYT #11200481.
References
1. Renard, F., Guedria, S., Palma, N. de & Vuillerme, N. Variability and reproducibility in deep learning for medical image segmentation. Scientific Reports 2020 10:1 10, 1–16 (2020).
2. Chen, C. et al. Deep Learning for Cardiac Image Segmentation: A Review. Frontiers in Cardiovascular Medicine 0, 25 (2020).
3. Rawat, W. & Wang, Z. Deep convolutional neural networks for image classification: A comprehensive review. Neural Computation 29, 2352–2449 (2017).
4. Bernard, O. et al. Deep Learning Techniques for Automatic MRI Cardiac Multi-Structures Segmentation and Diagnosis: Is the Problem Solved? IEEE Transactions on Medical Imaging 37, 2514–2525 (2018).
5. Painchaud, N. et al. Cardiac Segmentation With Strong Anatomical Guarantees. IEEE transactions on medical imaging 39, 3703–3713 (2020).
6. Parreño, M., Paredes, R. & Albiol, A. Deidentifying MRI Data Domain by Iterative Backpropagation. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) 12592 LNCS, 277–286 (2020).
7. Ronneberger, O., Fischer, P. & Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) 9351, 234–241 (2015).
8. Campello, V. M. et al. Multi-Centre, Multi-Vendor, and Multi-Disease Cardiac Segmentation: The M&Ms Challenge. IEEE Transactions on Medical Imaging (2021) doi:10.1109/TMI.2021.3090082.
9. E. Heiberg, J. Sjögren, M. Ugander, M. Carlsson, H. Engblom, and H. Arheden, Design and Validation of Segment – a Freely Available Software for Cardiovascular Image Analysis, BMC Medical Imaging, 10:1, 2010.
10. León, M. A., Salas, R., Uribe, S. & Sotelo, J. Repository: CARDIAC SEGMENTATION FRAMEWORK FOR CINE MRI. https://github.com/alejo101010/VENTSEG (2021).
11. Barreiro-Pérez, M. et al. Left ventricular global myocardial strain assessment comparing the reproducibility of four commercially available CMR-feature tracking algorithms. European radiology 28, 5137–5147 (2018).
Figures