4772
Cardiac MRF Using Rosette Trajectories for Simultaneous Myocardial T1, T2, and Proton Density Fat Fraction Mapping1Department of Radiology, University of Michigan, Ann Arbor, MI, United States, 2Department of Biomedical Engineering, University of Michigan, Ann Arbor, MI, United States
Synopsis
This work aims to extend rosette cardiac MRF to enable simultaneous myocardial T1, T2, and PDFF mapping from a single scan. The accuracy of rosette cMRF in T1, T2, and PDFF quantification is validated in the ISMRM/NIST system phantom and an in-house built fat fraction phantom, respectively. Studies in healthy subjects show that rosette cMRF potentially enables more accurate T1 and T2 measurements than spiral cMRF in the myocardium due to reduced fat contamination.
Introduction
The goal of this work is to extend cardiac MR Fingerprinting (cMRF) with a rosette-based readout for T1, T2, and fat fraction mapping. cMRF is a novel multi-parametric mapping method that was initially shown to enable simultaneous myocardial T1 and T2 quantification1. While many MRF approaches do not distinguish water from fat, elimination of the signal from fat may reduce mapping errors caused by water-fat partial volume effects. Moreover, quantitative proton density fat fraction (PDFF) mapping may provide additional value in diagnosing disease2,3. Previously, the Dixon method has been incorporated in the cMRF framework using multi-echo radial acquisitions to enable T1, T2, and PDFF quantification4. Previous work has demonstrated that water-fat separation can be achieved along with myocardial T1 and T2 mapping using cMRF with rosette trajectories5. In this work, rosette cMRF is extended to enable simultaneous myocardial T1, T2, and PDFF mapping from a single scan.Methods
A rosette trajectory with eight lobes and a readout duration of 7.7ms (Figure 1a) was designed to suppress signals at -220Hz (the main resonance frequency of fat at 1.5T). Simulation studies show that this trajectory suppresses 94.7% of the signal at -220Hz (Figure 1b). This readout trajectory was incorporated into a previously reported 15-heartbeat 5-segment ECG-triggered cMRF sequence structure5 with acquisition window ~250 ms at late diastole, resulting in a total of 390 highly undersampled images for pattern matching. The spatial resolution was 1.56×1.56mm2 and the slice thickness was 8mm in all phantom and in vivo experiments.All experiments were performed on a 1.5T scanner (Siemens Sola, Erlangen, Germany). Rosette cMRF data were collected in the T2 layer of the ISMRM/NIST MRI system phantom 6,7 and an in-house fat fraction phantom8. T1 and T2 values obtained using rosette cMRF were compared with gold standard values measured using inversion recovery and single-echo spin echo methods. Three-point Dixon GRE measurements with optimal echo times at 1.5T (1.9/3.4/4.9ms) were used as the gold-standard PDFF values. 14 healthy subjects were scanned after written informed consent in this IRB-approved study. Mid-ventricular level short axis slices in the heart were acquired using the proposed rosette cMRF sequence and the original spiral 15-heartbeat cMRF sequence.5 Conventional T1 and T2 mapping methods (MOLLI and T2-prepared bSSFP) were also performed in 10 of the subjects.
Undersampled water and fat images and a B0 map were generated using the rosette cMRF approach, as described in previous work5. T1 and T2 maps for both water and fat were generated by matching undersampled images to a dictionary that models the subject’s cardiac rhythm5 and includes corrections for slice profile, preparation pulse efficiency9, and B05. To generate quantitative PDFF maps, the 8-lobe trajectory was divided into 9 segments (Figure 1a). Data from each segment were projected onto a low-dimensional subspace of rank 6 derived from the singular value decomposition (SVD) of the dictionary. Subspace images corresponding to the first singular value from each of the 9 rosette segments served as multi-echo images and were used to generate a PDFF map using Hierarchical IDEAL10 with noise correction11.
ROIs in each vial in the phantom study, and on T1 and T2 maps in segments 7-12 of the standardized AHA model for in vivo measurements, were drawn manually. The mean and standard deviation in T1 and T2 values were calculated for each ROI, as well as over the entire myocardium in the in vivo maps. A student’s t-test was used to compare T1 and T2 measurements using different methods. Significant difference was considered with P<0.05.
Results
In the ISMRM/NIST system phantom, T1 and T2 measurements using rosette cMRF are in excellent agreement with the reference values (Figure 2). In the fat fraction phantom, water and fat specific T1 and T2 maps, proton density images, and the PDFF map generated by Hierarchical IDEAL using the rosette cMRF data themselves are shown in Figure 3a. Quantitative PDFF measurements using rosette cMRF agree well with 3-point Dixon GRE measurements (Figure 3b).Representative maps and images from one healthy subject are shown in Figure 4. The averaged T1 and T2 values of all subjects in each segment as well as in the entire myocardium are shown in Figure 5. Over the myocardium, spiral cMRF yielded similar T1 values (1004.7 ± 53.6 ms) compared with MOLLI (994.8 ± 18.8 ms) while rosette cMRF generated significantly higher T1 values (1081.8 ± 33.6 ms); both spiral and rosette cMRF yielded lower T2 values (spiral 37.3 ± 2.9 ms; rosette 40.4 ± 1.4 ms) compared with the conventional method (45.8 ± 2.4 ms), which is consistent with previous findings5. Averaged PDFF in the entire slice was 0.6% among all volunteers.
Discussion and Conclusions
This study shows that rosette cMRF can enable efficient cardiac tissue characterization through the simultaneous quantification of myocardial T1, T2, and PDFF. Note that in segment 7 (anterior wall) which is surrounded by epicardial fat, the difference in T1 measured by rosette cMRF and spiral cMRF is more pronounced than in other segments; spiral cMRF yielded higher T2 values than rosette cMRF, which is opposite to the trend in the other segments (Figure 5). Given that fat has T1 of 300~370 ms (lower than myocadium) and T2 of ~53ms (higher than myocardium) at 1.5T12, these measurements are possibly due to reduced fat contamination in rosette cMRF. Studies on cardiac patients with abnormal fat deposition will be performed in the future.Acknowledgements
This work is supported by NSF/CBET 1553441, NIH/NHLBI R01HL094557, and Siemens Healthineers (Erlangen, Germany).References
1. Hamilton, J. I. et al. MR fingerprinting for rapid quantification of myocardial T1, T2, and proton spin density. Magn. Reson. Med. 77, 1446–1458 (2017).
2. Farrelly, C., Shah, S., Davarpanah, A., Keeling, A. N. & Carr, J. C. ECG-gated multiecho Dixon fat-water separation in cardiac MRI: Advantages over conventional fat-saturated imaging. Am. J. Roentgenol. 199, (2012).
3. Kellman, P. et al. Multiecho dixon fat and water separation method for detecting fibrofatty infiltration in the myocardium. Magn. Reson. Med. 61, 215–221 (2008).
4. Jaubert, O. et al. Water–fat Dixon cardiac magnetic resonance fingerprinting. Magn. Reson. Med. 00, 1–17 (2019).
5. Liu, Y., Hamilton, J., Eck, B., Griswold, M. & Seiberlich, N. Myocardial T1 and T2 quantification and water–fat separation using cardiac MR fingerprinting with rosette trajectories at 3T and 1.5T. Magn. Reson. Med. (2021) doi:10.1002/mrm.28404.
6. Keenan, K. et al. Multi-site, multi-vendor comparison of T1 measurement using ISMRM/NIST system phantom. in Proc Int Soc Magn Reson Med. 24 3290 (2016).
7. Russek, S. et al. Characterization of NIST/ISMRM MRI system phantom. in Proc Int Soc Magn Reson Med. 20 2456 (2012).
8. Hines, C. D. G. et al. T1 independent, T2* corrected MRI with accurate spectral modeling for quantification of fat: Validation in a fat-water-SPIO phantom. J. Magn. Reson. Imaging 30, 1215–1222 (2009).
9. Hamilton, J. I. et al. Investigating and reducing the effects of confounding factors for robust T1 and T2 mapping with cardiac MR fingerprinting. Magn. Reson. Imaging 53, 40–51 (2018).
10. Tsao, J. & Jiang, Y. Hierarchical IDEAL: Fast, robust, and multiresolution separation of multiple chemical species from multiple echo times. Magn. Reson. Med. 70, 155–159 (2013).
11. Liu, C. Y., McKenzie, C. A., Yu, H., Brittain, J. H. & Reeder, S. B. Fat quantification with IDEAL gradient echo imaging: Correction of bias from T1 and noise. Magn. Reson. Med. 354–364 (2007) doi:10.1002/mrm.21301.
12. Rakow-Penner, R., Daniel, B., Yu, H., Sawyer-Glover, A. & Glover, G. H. Relaxation times of breast tissue at 1.5T and 3T measured using IDEAL. J. Magn. Reson. Imaging 23, 87–91 (2006).
Figures
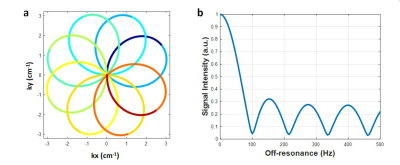
Figure 1. (a) The rosette trajectory used in this work, where the segments used to generate images at different echo times are indicated by different colors. (b) Spectral response of the trajectory. Fat signals at -220Hz are suppressed to 5.3%.
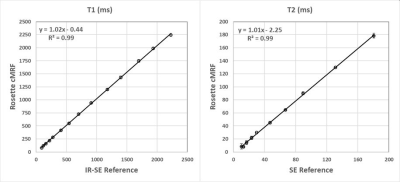
Figure 2. T1 and T2 measurements in the ISMRM/NIST system phantom using rosette cMRF compared with reference values.
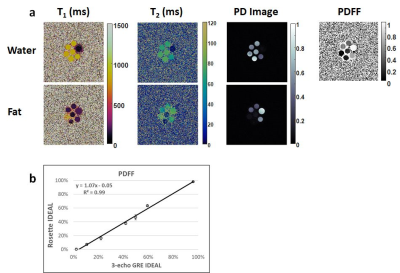
Figure 3. (a) Water and fat specific T1 and T2 maps, proton density (PD) images, and the PDFF map generated by Hierarchical IDEAL using the rosette cMRF data. (b) PDFF measurements in the fat fraction phantom using rosette cMRF compared with reference values.
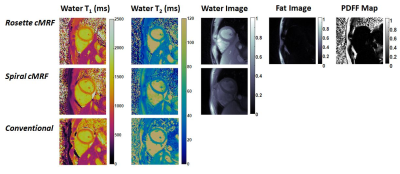
Figure 4. Representative T1 map, T2 map, water image, fat image, and PDFF maps in a healthy subject acquired using rosette cMRF as well as comparison with T1 and T2 maps measured by spiral cMRF and conventional methods. The field-of-view has been cropped to 150×150mm2 to better visualize the heart.
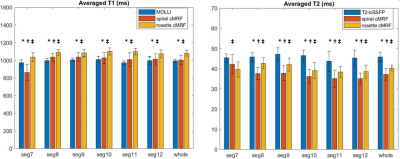
Figure 5. T1 and T2 values in 14 healthy subjects measured using conventional methods, spiral cMRF, and rosette cMRF. Measurements in segment 7 to 12 as well as over the entire myocardium are shown.
* Significant difference between spiral and rosette cMRF.
† Significant difference between conventional method and spiral cMRF.
‡ Significant difference between conventional method and rosette cMRF.