4756
A deep learning dipole inversion method for QSM of arbitrary head orientation and image resolution1School of ITEE, the University of Queensland, Brisbane, Australia
Synopsis
Due to the intrinsic data-driven property, many existing deep learning QSM methods can only be applied to local field maps with FOV orientation and image resolution consistent with the training data. This work proposes a novel and robust deep learning approach to reconstruct QSM of arbitrary head orientation and image resolution. Experiments are conducted on both simulated and in vivo human brain data to verify the proposed approach.
Introduction
Quantitative susceptibility mapping (QSM) measures the magnetic susceptibility property of biological tissue by solving an ill-posed problem based on the tissue field map. Most existing deep learning QSM models are trained on pure-axial acquisition data with 1 mm isotropic resolution. Despite the improved QSM results these approaches [1-3] have demonstrated, they fail to perform optimally when the orientation or resolution of the local field acquisition varies from its training dataset. This study proposes a novel deep learning solution that accommodates local field maps with oblique field-of-views (FOVs) and arbitrary image resolutions by performing local field affine transformation and image deblurring in a single U-net 4.Methods
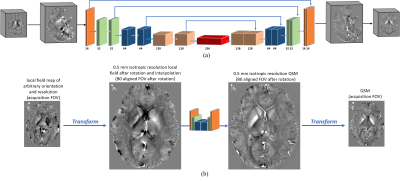
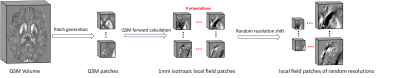
As demonstrated in Fig. 1, a local field in arbitrary orientation and resolution is first resampled, rotated, and interpolated to pure axial acquisition in 0.5mm isotropic resolution by following affine transformation rules. This transformed local field map is then fed into a conventional U-net, where a susceptibility map is computed. Finally, another inverse affine transformation is performed on the calculated susceptibility map, by which the original orientation and resolution are restored.The model was trained on patches due to GPU memory constrain. As illustrated in Fig. 2, a total of 15,360 small patches (483) were cropped from 96 full-sized 1 mm isotropic brain QSM images (144x192x128). For each patch, eight differently obliqued (ranging from 15° to 45° tilted to the main field direction) and one pure axial local field patches were simulated via the forward dipole convolution. These 1mm isotropic local field patches from nine head orientations were resampled to image resolutions randomly chosen from [0.5, 0.8, 1.0, 1.2, 1.5, 1.8, 2.0] (mm) for each dimension. The same forward transformation was also applied to generate the 0.5mm isotropic pure-axial QSM patches used for a second training loss. For comparison, a conventional U-net is also trained with only pure axial local field patches in 1 mm isotropic resolution, as commonly adopted in previous deep learning QSM methods [1, 2].
The proposed U-net, including all affine transformation operations, is implemented using Pytorch, thus computed gradients can be thoroughly propagated forward and backward during optimization. The Adam optimizer and smooth L1 loss were adopted for network training. To align the patch size with different resolutions and ensure the rotated result is in the current image view, each patch is padded with 48 empty voxels, which leads to a relatively large patch size of 1443. It took 60 hours to train for 80 epochs using two Tesla V100 GPUs with a mini-batch size of 12.
Results
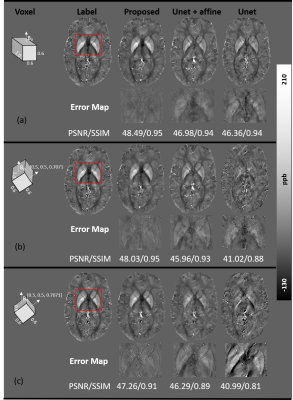
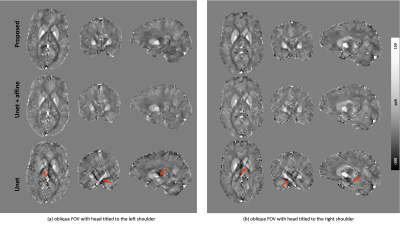
Fig. 3 compares the proposed pipeline with a conventional U-net on simulated local field maps with two acquisition angles (i.e., large-angle (Hlab = [0.5, 0.5, 0.707]) and pure axial (Hlab = [0, 0, 1]) and two image resolutions (i.e., 0.6 mm isotropic and 0.6x0.6x1 mm3). Conventional U-net failed to reconstruct QSM from either resolution whenever the FOV is oblique (Fig. 3 b and c). The U-net results improve with added affine transformations (i.e., denoted as U-net+affine in Fig. 3), however, noticeable susceptibility underestimation is still observed compared to the proposed method.Results from in vivo experiments of two oblique orientations with 0.6 mm isotropic resolution acquired at a 7T MRI system are shown in Fig. 4. Consistent with the simulation results, the conventional U-net led to substantial streaking artifacts and apparent errors (red arrows in Fig. 4) due to dipole inversion effects observable in all three orthogonal planes. Meanwhile, our proposed method successfully reconstructed QSM without reducing susceptibility contrast as in the standard U-net+affine results.
Discussion
We develop a deep learning QSM method for arbitrary image orientation and resolution by integrating affine transformations with U-net in an All-in-One pipeline. Affine transformation resolves orientation and resolution mismatches, and the All-in-One U-net compensates for the introduced image blurring and susceptibility under-estimation. Thus, the new method improves the generalizability of the neural network for various input data.Conclusion
This work shows that affine transformations of the input data can improve the predictions from deep neural network for QSM dipole inversion of arbitrary image orientation and resolutions. By training the proposed network with various orientations and resolutions, a single U-net can simultaneously perform image deblurring while computing dipole inversion.Acknowledgements
Hongfu Sun acknowledges support from the Australian Research Council (DE210101297).References
1. Yoon J, Gong E, Chatnuntawech I, et al. Quantitative susceptibility mapping using deep neural network: QSMnet[J]. Neuroimage, 2018, 179: 199-206.
2. Bollmann S, Rasmussen K G B, Kristensen M, et al. DeepQSM-using deep learning to solve the dipole inversion for quantitative susceptibility mapping[J]. Neuroimage, 2019, 195: 373-383.
3. Gao Y, Zhu X, Moffat B A, et al. xQSM: quantitative susceptibility mapping with octave convolutional and noise‐regularized neural networks[J]. NMR in Biomedicine, 2021, 34(3): e4461.
4. Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation[C]//International Conference on Medical image computing and computer-assisted intervention. Springer, Cham, 2015: 234-241.
5. Li W, Wang N, Yu F, et al. A method for estimating and removing streaking artifacts in quantitative susceptibility mapping[J]. Neuroimage, 2015, 108: 111-122.
Figures