4706
MGMT Promoter Methylation Prediction by End-to-end Evidential-Efficient Net1Zhejiang Univerisity, Hangzhou, China, 2The First Affiliated Hospital of Guangzhou Medical University, Guangzhou, China, 3Brigham and Women's Hospital,Harvard Medical School, Boston, MA, United States
Synopsis
Malignant brain tumor affects a large number of patients and often have poor prognosis and low response to therapeutics. An indicator of the progress of brain tumor and its response to treatment is the DNA repair protein, O6-methylguanine-DNA methyltransferase (MGMT). As such, accurate assessment of MGMT is of great clinical significance. Biospy is not only invasive but also has the risk of undersampling the tumorous tissue. We presented a novel deep learning model that uses multi-MRI modalities to assess the expression of MGMT in glioblastoma (GBM) patients. Results showed that the model can achieve good performance.
Introduction
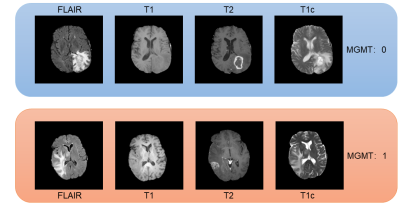
GBM is the most popular malignant brain tumor and has a poor prognosis. Most GBM patients have a survival of less than one year. MGMT promotor methylation has been shown to be a prognostic factor that affects the length of survival of GBM patients and indicates how likely a GBM may respond to alkylating chemotherapy. As such, assessing the status of MGMT in GBM patients is of significant clinical importance. Current practice requires biopsy and genetic analysis, a process that, in addition to be invasive, can take days to complete. It is therefore beneficial to develop methods for predicting MGMT expression using brain imaging1 (Figure1). We presented a deep learning approach based on end-to-end Evidential-Efficient-Net (EEN) to extract radiomics features from brain MRI and predict MGMT expression.Methods
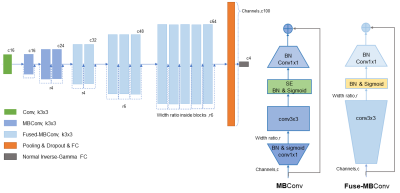
The input data to our deep learning model includes four MRI modalities, namely, T1, T2, FLAIR, and T1c (T1 contract enhanced). We extracted 582 patient cases from BraTS2021 database2. These cases were selected because they had the four MRI scans and known MGMT groundtruth. We randomly separated the cases into a training set of 466 patients and a test set of 116 patients. All the MRIs were resized to 256 by 256 pixels in axial scans. For some scans that were generated in the coronal and sagittal directions, we transformed the scans to the axial direction.The core component of our method is the EfficientNetV23, which, in our method, was scaled down to a tiny net that we named as EffNetV2-T. The EffNetV2-T preserved the MBConv and Fused-MBConv block in the original EfficientNet in the architecture searching space. We adjusted the channels and the SE lay inside the blocks to accommodate the four MRI modalities. The original EfficientNetV2-S proves too deep for our task, it would cause seriously overfitting and reach a low bottleneck. To address these problem, besides scale down the network, we introduced dropout layers and adjust regularization as well. To achieve high performance, we incorporated the idea of evidential deep learning (EDL) 4 into our model. EDL is a novel method for training non-Bayesian neuron networks to estimate a continuous target as well as its associated evidence to learn both aleatoric and epistemic uncertainty. We employed an evidential layer as the last layer of our model. The evidential layer transfers the image features to a Normal Inverse-Gamma (NIG) distribution:
$$p(\mu,\sigma^2|\gamma,\nu,\alpha,\beta)=\frac{\beta^\alpha\ \sqrt v}{\mathrm{\Gamma}(\alpha)\ \sqrt{2\pi\sigma^2}} (\frac{ 1}{\sigma^2\ })^{\alpha+1}\ \exp \left\{- \frac {2\beta+\nu(\gamma-\mu)^2}{2\sigma^2\ } \right\}$$
With an evidential loss function: $$$L_i\ (w)=L_i^{NLL}\ (w)+\lambda L_i^R\ (w)$$$, in which negative log likelihood(NLL) is defined for sample as $$$L_i^{NLL} = -log [p(y_i|m)]$$$ and the evidence regularizer that scaled by a regularization coefficient , $$$L_i^R(w)= |y_i-\gamma |·(2\nu + \alpha)$$$.
Our model formulates learning as an evidence acquisition process, in which, every training example adds information to maximizing the model fit and minimizing evidence on errors. To balance the uncertainty inflation and the accuracy, we use maximum rate and ε to constraint the fluctuation of λ. we use Adam optimizer, and set the original learning rate as 0.01. Our model reaches the best score within 50 epoch’s training, and shows little signs of overfitting in five-cross validation. In other words, our model used the modified EffNetV2-T for feature extraction and evidential regression for classifying the status of MGMT. We name the final model as Evidential-EffNet (Figure 2).
Results
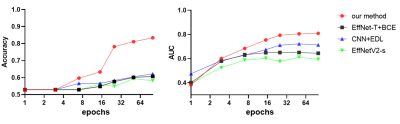
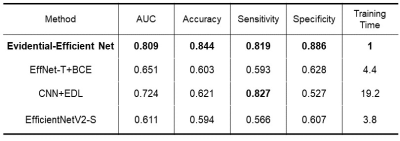
We used the area under the receiver operating characteristics curve (AUC) to measure the performance of the proposed model. Our results on the test set showed that the model obtained an accuracy of 84.4% on average and an AUC of 0.809, indicating its good performance. We also compared our model to others, which shows that our model has the best performance. (Figure 3 and Table 1)Discussion and Conclusions
We presented a novel model that integrates EfficientNetV2 and EDL to predict MGMT expression using brain MRI. We compared our model to the EffNetV2-T with Binary Cross Entropy as the loss function and found that our model can train faster and obtain better results. We also compared our model to an EffNet-like convolutional neural network without network architecture searching. The result showed that our model can train faster and obtain better result. Our model also compared favorably with EffNet-B0-4, which uses only MBConv blocks as our model has a faster training speed and higsher performance.Acknowledgements
No acknowledgement found.References
1. Levner, Ilya, et al. "Predicting MGMT methylation status of glioblastomas from MRI texture." International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Berlin, Heidelberg, 2009.
2. Baid, Ujjwal, et al. "The rsna-asnr-miccai brats 2021 benchmark on brain tumor segmentation and radiogenomic classification." arXiv preprint arXiv:2107.02314 (2021).
3. Tan, Mingxing, and Quoc V. Le. "Efficientnetv2: Smaller models and faster training." arXiv preprint arXiv:2104.00298 (2021).
4. Amini, Alexander, et al. "Deep evidential regression." Advances in Neural Information Processing Systems 33 (2020).