4682
Reconstructing T2 Maps of the Brain from Highly Sparse k-Space Data with Generalized Series-Assisted Deep Learning1School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China, 2Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 3Department of Electrical and Computer Engineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 4Radiology Department, The Fifth People's Hospital of Shanghai, Shanghai, China
Synopsis
Quantitative T2 maps of brain tissues are useful for diagnosis and characterization of a number of diseases, including neurodegenerative disorders. This work presents a new learning-based method for the reconstruction of T2 maps from highly sparse k-space data acquired in accelerated T2-mapping experiments. The proposed method synergistically integrates generalized series modeling with deep learning, which effectively captures the underlying signal structures of T2-weighted images with variable TEs and a priori information from prior scans (e.g., data from the Human Connectome Project). The proposed method has been validated using experimental data, producing improved brain T2 maps over the state-of-the-art methods.
Introduction
The past few years have seen a surge of interest in using deep learning-based methods to reconstruct T2 maps from highly sparse imaging data.1-5 A practical challenge for deep learning-based T2 mapping lies in the lack of multi-TE experimental data with ground truth required for network training. An effective method was recently proposed to address this issue, which involves the use of single-TE image priors, as well as generalized series (GS) and sparse modeling.6 This work extends this method by integrating GS modeling with deep learning (DL). This novel integration exploits the complementary information provided by the GS model and deep network to substantially improve the reconstruction quality. In addition, the proposed method enables the use of synthetic data for network training because the deep network was trained only to compensate the GS model. The proposed method has been validated using experimental data, producing more accurate brain T2 maps as compared with the state-of-the-art methods.Method
Following our recent study,6 we decomposed the desired image functions of different TEs as:$$\rho(\boldsymbol{x}, t)=\sum_{m=-M}^{M} \alpha_{m}(t) \rho_{\mathrm{ref}}(\boldsymbol{x}, t) e^{-i 2 \pi m \Delta \boldsymbol{k} \cdot \boldsymbol{x}}+\rho_{\mathrm{s}}(\boldsymbol{x}, t)$$
where $$$t=\mathrm{TE}_{1}, \mathrm{TE}_{2}, \ldots, \mathrm{TE}_{n}$$$;$$$\rho_{\text {ref }}(\boldsymbol{x}, t)$$$ represents the GS reference image for absorbing the anatomical information;$$$\rho_{\mathrm{s}}(\boldsymbol{x}, t)$$$ denotes the sparse component for capturing subject-dependent novel features. In previous study, $$$\rho_{\text {ref }}(\boldsymbol{x}, t)$$$ was obtained using a deep learning-based image translation scheme that generated reference images for different TEs from the companion T1-weighted image and $$$\rho_{\mathrm{s}}(\boldsymbol{x}, t)$$$ was determined by solving an $$$L_{1}$$$ norm-based compressed sensing reconstruction problem.
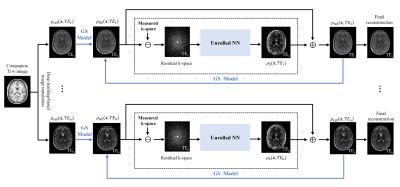
In this work, we further generalized this reconstruction framework by synergistically integrating GS modeling with deep learning. The proposed method has three novel features to improve reconstruction performance. First, instead of using a hand-crafted sparsity-promoting regularizer for $$$\rho_{\mathrm{s}}(\boldsymbol{x}, t)$$$, we learned its optimal functional form through an unrolled network. This data-driven regularizer has been demonstrated to be more effective in constraining the recovery of image features.7-10Second, instead of fixing $$$\rho_{\text {ref }}(\boldsymbol{x}, t)$$$, we iteratively updated them by including the recovered novel features produced by the network. In this way, the deep network only serves to compensate the spatial basis functions in the GS model, and consequently, it can be trained using simulated data. This feature is especially desirable for T2 mapping where only a small number of multi-TE training data are currently available. Finally, we included the output of the GS reconstruction into the deep network as a conditional prior to improve its accuracy and robustness. The overall pipeline of the proposed method is shown in Figure 1.
Specifically, in the first iteration, we generated $$$\rho_{\text {ref }}(\boldsymbol{x}, t)$$$ and determined the GS component $$$\hat{\rho}_{\mathrm{GS}}(\boldsymbol{x}, t)$$$ as was done in6. Then, we used a deep network to reconstruct $$$\rho_{\mathrm{s}}(\boldsymbol{x}, t)$$$ which unrolled the gradient descent algorithm for solving the following reconstruction problem:
$$\hat{\rho}_{\mathrm{s}}=\arg \min _{\rho_{\mathrm{s}}} \frac{1}{2}\left\|d-F_{\Omega}\left(\hat{\rho}_{\mathrm{GS}}+\rho_{\mathrm{s}}\right)\right\|^{2}+R\left(\rho_{\mathrm{s}} ; \hat{\rho}_{\mathrm{GS}}\right)$$
As described in7, this unrolled network equivalently learned the optimal regularized functional $$$R(\cdot)$$$ for recovery of $$$\hat{\rho}_{\mathrm{s}}$$$. Note differently from7, we conditioned the network input on $$$\hat{\rho}_{\mathrm{GS}}$$$ so that the prior information therein was incorporated to further improve and stabilize the network. Synthetic data were used to train the network, which were generated by transferring the T2-weighted images of Human Connectome Project11 to T2-weighted images for different TEs through histogram matching.
After $$$\hat{\rho}_{\mathrm{S}}(\boldsymbol{x}, t)$$$ was determined, we updated the reference image as $$$\rho_{\mathrm{ref}}(\boldsymbol{x}, t)=\hat{\rho}_{\mathrm{GS}}(\boldsymbol{x}, t)+\hat{\rho}_{\mathrm{s}}(\boldsymbol{x}, t)$$$ and generated a set of new spatial basis functions for the GS model. The updated spatial bases matched better with the data, thus making the GS model-based reconstruction more accurate and the corresponding residual feature sparser. As a result, the unrolled network more effectively used the imaging data for signal recovery. By alternatively updating the GS model and unrolled network until convergence, we obtained the final reconstruction.
Results
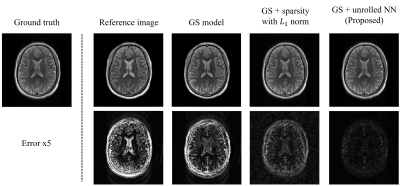
The proposed method was evaluated using experimental data obtained from the hospital.Figure 2 shows the role of the GS model and the unrolled NN in producing the final reconstruction. As can be seen, the low-frequency discrepancies between the reference image and the ground truth were compensated by the GS model. The sparse image features were further recovered by the unrolled NN. Compared to handcrafted sparse constraints, the learned regularization term better recovered the sparse image features.
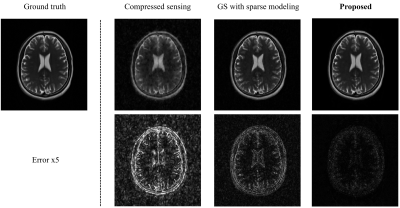
Figure 3 compares the reconstructed T2-weighted images using the proposed method with those from two existing state-of-the-art methods. As shown, the proposed method produced significantly improved results.
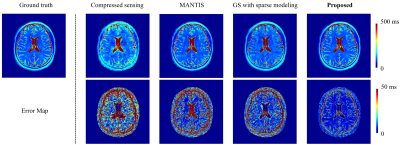
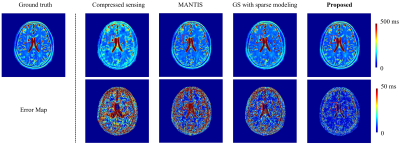
Figure 4 compares the accuracy of the T2 maps obtained using the proposed method with those of three existing state-of-the-art methods. Again, the proposed method produced much better T2 maps.
Figure 5 shows a comparison of the proposed method with three state-of-the-art methods in recovering T2 maps from data acquired from a patient. As can be seen, the proposed method was able to detect the lesion clearly.
Conclusions
We presented a new method to reconstruct brain T2 maps from highly sparse imaging data. The proposed method synergistically integrates GS modeling with deep learning for substantially improved reconstruction accuracy. Experimental results demonstrate its superior performance over state-of-the-art methods. Our method may prove useful for various MRI studies that involve quantitative T2 mapping.Acknowledgements
No acknowledgement found.References
[1] Liu F, Feng L, Kijowski R. MANTIS: Model‐Augmented Neural neTwork with Incoherent k‐space Sampling for efficient MR parameter mapping. Magnetic resonance in medicine. 2019; 82(1): 174-188.
[2] Liu F, Kijowski R, Feng L, et al. High-performance rapid MR parameter mapping using model-based deep adversarial learning. Magnetic Resonance Imaging. 2020; 74: 152-160.
[3] Cai C, Wang C, Zeng Y, et al. Single‐shot T2 mapping using overlapping‐echo detachment planar imaging and a deep convolutional neural network. Magnetic resonance in medicine. 2018; 80(5): 2202-2214.
[4] Fang Z, Chen Y, Liu M, et al. Deep learning for fast and spatially constrained tissue quantification from highly accelerated data in magnetic resonance fingerprinting. IEEE transactions on medical imaging. 2019; 38(10): 2364-2374.
[5] Cohen O, Zhu B, Rosen M S. MR fingerprinting deep reconstruction network (DRONE). Magnetic resonance in medicine. 2018; 80(3): 885-894.
[6] Meng Z, Guo R, Li Y, et al. Accelerating T2 mapping of the brain by integrating deep learning priors with low‐rank and sparse modeling. Magnetic Resonance in Medicine. 2021; 85(3): 1455-1467.
[7] Hammernik K, Klatzer T, Kobler E, et al. Learning a variational network for reconstruction of accelerated MRI data. Magnetic resonance in medicine. 2018; 79(6): 3055-3071.
[8] Yang Y, Sun J, Li H, et al. ADMM-CSNet: A deep learning approach for image compressive sensing. IEEE transactions on pattern analysis and machine intelligence. 2018; 42(3): 521-538.
[9] Aggarwal H K, Mani M P, Jacob M. MoDL: Model-based deep learning architecture for inverse problems. IEEE transactions on medical imaging. 2018; 38(2): 394-405.
[10] Schlemper J, Caballero J, Hajnal J V, et al. A deep cascade of convolutional neural networks for dynamic MR image reconstruction. IEEE transactions on Medical Imaging. 2017; 37(2): 491-503.
[11] Van Essen DC, Smith SM, Barch DM, et al. The WU-Minn Human Connectome Project: an overview. Neuroimage. 2013;80:62-79.
Figures