4616
Accelerated 3D PDWI in knee imaging: Quality and efficiency of a deep learning-based Compressed SENSE reconstruction1Radiology, The First Hospital of Jilin University, Changchun, China, 2The First Hospital of Jilin University, Changchun, China, 3College of Electronic Science and Engineering, Jilin University, Changchun, China, 4Philips Healthcare, Beijing, China
Synopsis
The use of three dimensional (3D) volumetric acquisition in clinical settings has been limited due to long scan time. A deep learning-based reconstruction algorithm allows shortening of scan time and provide comparable overall image quality when compared with standard sequences. Adaptive-CS-Net, a deep neural network previously introduced at the 2019 fast MRI challenge, was expanded and presented here as a Compressed-SENSE Artificial Intelligence (CS-AI) reconstruction. The purpose of the study is to determine the feasibility of 3D PDWI accelerated with CS-AI for evaluating the knee image quality and compared with SENSE and standard Compressed-SENSE.
Introduction
Knee pain affects approximately 25 % of adults, limits function and mobility, and impairs quality of life (1). MRI offers an accurate evaluation of musculoskeletal soft tissues (meniscus, ligament, tendon, and muscle), as well as occult bone injuries. Despite the use of parallel imaging, the available three-dimensional (3D) sequences require more than 5 minutes per scan in clinical routine. However, 3D volumetric acquisition could provide higher spatial resolution and decrease partial volume effects. Longer scan time makes 3D scan sequences more prone to motion artifacts and hence may degrade image quality. Integrating artificial intelligence (AI) into the MRI reconstruction has attracted much attention. At the 2019 fast MRI challenge, a novel deep neural network was introduced as Adaptive-CS-Net and showed superior performance for reconstructing MRI images from highly under sampled k-space data (2). The Adaptive-CS-Net was expanded to multiple contrasts and anatomical areas and is presented here as Compressed SENSE AI (CS-AI) reconstruction (3). The purpose of this study was to acquire highly accelerated knee MRI using the CS-AI reconstruction and compare the image quality with the conventional method, SENSE (S), Compressed-SENSE (CS) and CS-AI with different acceleration factors (AF).Methods
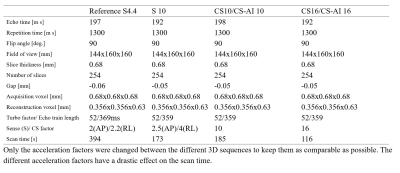
The study was approved by the local IRB, and written informed consent was obtained from all subjects. A total of 23 volunteers were examined on a 3.0T whole-body clinical system (Ingenia Elition, Philips Healthcare) using a dedicated knee coil, all receiving a SENSE with AF 10 (S 10), CS with AF 10 and 16 (CS 10 and CS 16) and time-equivalent CS-AI with AF 10 and 16 (CS-AI 10 and CS-AI 16) fat suppressed 3D proton density weighted imaging (PDWI) sequence. The commercially available fat suppressed 3D PDWI scan was acquired with parallel imaging acceleration of 4.4 (S 2 × 2.2) as reference. The sequence parameters are summarized in Table 1. The CS-AI model used in this study is the extension of the previously introduced AI-based reconstruction algorithm, Adaptive-CS-Net (4, 5). In CS-AI, the iterative optimization procedure in the C-SENSE reconstruction chain is unrolled for a fixed number of reconstruction blocks. Each block consists mainly of U-Net-like architecture, which performs as a denoiser. Quantitative image analysis was performed by three radiologists with more than 5 years of experience. ROIs were placed on bone (distal femur), muscle (gastrocnemius muscle), meniscus (the anterior horn of the lateral meniscus), anterior cruciate ligament (ACL) and synovial fluid. Based on the ROIs, Signal-to-noise ratio (SNR) and contrast-to-noise ratio (CNR) were analyzed for objective evaluation. All numerical values were reported as the mean ± SD. We used a paired t-test to compare the SNR, and CNR with the CS, CS-AI and reference SENSE sequences. P values< 0.05 were considered significant.Results
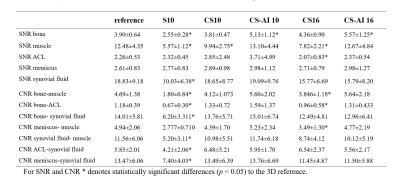
The 3D PDWI sequence was accelerated using two different acceleration factors. Scan time decreased with increasing AF (reference S4.4: 394s, S/CS/CS-AI10: 185s, CS/CS-AI16: 116s). The quantitative image analysis results for the different knee anatomical structures are shown in Table 2. The SNR and CNR values of S10 for most structures were significantly reduced to reference sequences and the image quality became very noisy. The SNR of CS10 and CS16 in some anatomical areas (muscle and ACL) were inferior to the reference sequence (all P <0.05), and the CNR (bone-muscle, bone-ACL, meniscus-muscle, ACL-meniscus) of CS 16 sequence were significantly reduced compared to the reference sequence. However, the SNR of bone area for CS-AI10 and CSAI-16 were even significantly higher than the reference sequence (P<0.05), and no difference was found for the SNR and CNR of other anatomical locations in any of the analyzed CS-AI sequences. Figure 1 showed image quality of CS,CS-AI is comparable to that of the reference method.Discussion
The CS-AI images demonstrated high denoising performance. In both acceleration factors of 10 and 16, the CS-AI with shorter scan time showed image quality is comparable to or better than that of the reference method. Both the reference and CS-AI showed good image quality, but the CS-AI demonstrated a better depiction of small structures such as cartilage in knee MRI and produced visually sharper images compared to SENSE and Compressed SENSE. The S10 and CS16 images became very noisy and were not suitable for effective clinical applications. The above results suggest that the scan acceleration with the noise reduction by CS-AI reconstruction may be useful for getting a high-quality image to improve diagnostic confident, and it is essential to enable this new technology to be implemented in clinical routine.Conclusion
Deep learning-based reconstruction for cross-sectional imaging may reduce image noise, improve image quality and potentially decrease acquisition time.Acknowledgements
None.References
1. Taylor NJRdcoNA. Nonsurgical Management of Osteoarthritis Knee Pain in the Older Adult: An Update. 2018;44(3):513-524.
2. Knoll F, Zbontar J, Sriram A, Muckley M, Bruno M, Defazio A, Parente M, Geras K, Katsnelson J, Chandarana H, Zhang Z, Drozdzalv M, Romero A, Rabbat M, Vincent P, Pinkerton J, Wang D, Yakubova N, Owens E, Zitnick C, Recht M, Sodickson D, Lui YJRAi. fastMRI: A Publicly Available Raw k-Space and DICOM Dataset of Knee Images for Accelerated MR Image Reconstruction Using Machine Learning. 2020;2(1):e190007.
3. Hammernik K, Klatzer T, Kobler E, Recht M, Sodickson D, Pock T, Knoll FJMrim. Learning a variational network for reconstruction of accelerated MRI data. 2018;79(6):3055-3071.
4. Pezzotti N, Yousefi S, Elmahdy MS, Van Gemert JHF, Schuelke C, Doneva M, Nielsen T, Kastryulin S, Lelieveldt BPF, Van Osch MJP, De Weerdt E, Staring M. An Adaptive Intelligence Algorithm for Undersampled Knee MRI Reconstruction. Ieee Access 2020;8:204825-204838.
5. Pezzotti N, de Weerdt E, Yousefi S, Elmahdy MS, van Gemert J, Schülke C, Doneva M, Nielsen T, Kastryulin S, Lelieveldt BPF, van Osch MJP, Staring MJae-p. Adaptive-CS-Net: FastMRI with Adaptive Intelligence. 2019. p arXiv:1912.12259.
Figures