4481
Intracellular Iron Chelation affects Cellular Metabolism in Triple Negative Breast Cancer Cells: Comparison between Human and Murine Models1Medical Physics, Memorial Sloan-Kettering Cancer Center, New York, NY, United States, 2Medical Physics, Memorial Sloan Kettering Cancer Center, New York, NY, United States, 3Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, NY, United States, 4Molecular Pharmacology Program, Memorial Sloan Kettering Cancer Center, New York, NY, United States, 5Weill Cornell Medical College, Cornell University, New York, NY, United States
Synopsis
We investigated the effect of Deferiprone, an iron chelator clinically used for non-cancer related diseases, on cellular metabolism and the impairment of cell growth in the human MDA-MB-231 and murine 4T1 triple-negative breast cancer models. By comparing the findings obtained on both cell lines through 13C MRS monitoring with those related to mitochondrial respiration, we aimed to better understand the metabolic mechanism driving the changes that follow Deferiprone exposure. A stronger effect of DFP exposure was observed in human MDA-MB-231 cells than murine 4T1 cells.
Introduction
Triple-negative breast cancers (TNBC) are aggressive tumours associated with poor prognosis and lacking in effective therapies.1 Cellular iron is essential for cell proliferation2 and vitality and its cellular level is often related to cancer progression.3,4 Here, we investigated the effect of Deferiprone (DFP), an iron chelator clinically used for non-cancer related diseases, on two TNBC cell lines with the goal to better understand the mechanism underpinning the metabolic changes following DFP exposure. Metabolism of both cells lines was monitored over time using an MR-compatible cell bioreactor, and the changes, following DFP exposure, were compared between both cell lines. Findings on quantitative mitochondrial parameters were investigated for both cell lines and compared with 13C MRS results. The effect of DFP on cell migration was studied in relation to changes of cell doubling times due to DFP treatment.Methods
Human MDA-MB-231 and murine 4T1 cells were grown in high glucose Dulbecco’s Modified Essential (DME) medium, supplemented with 2 mM glutamine, 10% fetal bovine serum, 100 U/ml penicillin and 100 µg/ml streptomycin at 37 ⁰C in a 5% CO2 humidified atmosphere. For the MR bioreactor studies, 6×106 MDA-MB-231 cells or 3×106 4T1 cells were seeded on microcarriers (0.5 ml; 125-212 μm diameter; PP3772, Corning, New York, NY) in six deep-form bacteriological Petri dishes until reaching approximately 70% confluence. Live cells were studied in an MR-compatible bioreactor5 on a Bruker AVANCE III 11.7T MRI system (Bruker BioSpin, Billerica, MA) equipped with a 10-mm multi-nuclear broad-band probe, tunable to 1H and 31P/13C nuclei. For each cell line, three independent experiments were performed in regular medium conditions (CTRL) and three in medium containing 100 µM DFP (DFP-treated). After 1 h setup, the unlabelled medium was replaced with medium containing 25 mM of 99% D-Glucose-1-13C to monitor the perfused cells by 13C-MRS. For each experiment, 1H-decoupled, Nuclear Overhauser Effect-enhanced 13C MR spectra (TR = 1200 s, 45° flip angle, 30 kHz sweep width, 8 k points, 1000 averages, 20 min acquisition time) were acquired repeatedly every six hours interspersed by 31P MRS for a total of 30 h. Quantification of MR spectral peaks was performed in the time domain using AMARES (jMRUI v6.0). Metabolites were normalized to the b-NTP signal of the initial 31P MR spectrum (bNTPinit).2,5 A two-way ANOVA followed by Tukey’s post hoc test was used to compare metabolite 13C-labelling rates. The cell doubling times were determined for untreated and DFP-treated MDA-MB-231 and 4T1 cells respectively. The effect of DFP on the migration of confluent MDA-MB-231 and 4T1 cells was assessed by wound healing assay.2 The mitochondrial response of both cell lines to DFP exposure was assessed from the oxygen consumption rates (OCRs) using the Seahorse XF Cell Mito Stress Test Kit on the Seahorse XFe96 flux analyser (Agilent, Santa Clara, CA).Results
Figure 1 shows the differences between representative 13C MR spectra of untreated (Fig. 1A) and DFP-treated (Fig.1B) MDA-MB-231 cells, both acquired at the start (1 h) and end (31 h) of the perfusion experiment. The quantitative evaluation of 13C spectral metabolites (Fig. 2) showed higher glucose consumption in DFP-treated MDA-MB-231 cells than DFP-treated 4T1 cells reaching significant levels at 6 and 24 hours of treatment (Fig. 2B). In addition, glutamate and lactate synthesis showed higher levels in DFP-treated MDA-MB-231 cells compared with DFP-treated 4T1 cells, reaching statistical significance at 24 hours (Fig. 2B). The doubling time of both cell lines was significantly increased with the treatment (Fig. 3). Wound healing was significantly reduced in DFP-treated 4T1 cells at 24 and 36 h exposure, delaying wound closure to about 48 h compared to the 36 h in untreated and 50 µM DFP-treated 4T1 cells (Fig. 4B). Conversely, cell migration was higher in DFP-treated than untreated MDA-MB-231 cells (Fig. 4A) reaching significance at about 24 hours while at later time points control cells catch up in the wound closure. Conversely, cell migration was higher in DFP-treated cells than in untreated MDA-MB-231 cells (Fig. 4A) reaching significance at about 24 hours, while control cells reached wound closure at the later time points. These findings indicate that wound closure is most affected by DFP exposure when at least 1 cell doubling time has passed. In both TNBC cell lines, DFP exposure decreased basal respiration, ATP production, maximal respiration, spare respiratory capacity (SRC), and % spare respiratory capacity (% SRC).Discussion
Our findings on 13C metabolic changes due to DFP treatment (Fig. 2) along with those related to the mitochondrial parameters (Fig. 5B) suggest that human MDA-MB-231 cells are more affected by the DFP treatment than the murine TNBC 4T1 cells. In comparison to MDA-MB-231 cells, this might be due to an enhanced reliance of 4T1 cells on glycolytic energy metabolism and/or its higher plasticity to switch between glycolytic and mitochondrial metabolism, as previously observed.2Conclusion
This study shows that DFP exposure to TNBC cell models has a stronger effect on the human MDA-MB-231 cells than on the murine 4T1 cells, as confirmed by 13C metabolic results and mitochondrial respiration findings.Acknowledgements
This work was support by grants W81ZWH-17-1-0525 (JAK) and P30CA008748 (Cancer Center Support Grant). We thank Dr. RV Simões for helpful discussions and the Molecular Cytology Core Facility for their assistance with the wound healing assay.
References
1 Collignon J, Lousberg L, Schroeder H et al. Triple-negative breast cancer: treatment challenges and solutions. Breast Cancer 2016, 8:93-107.
2 Simoes RV, Veeraperumal S, Serganova IS, et al. Inhibition of prostate cancer proliferation by Deferiprone. NMR Biomed. 2017;30(6).
3. Wang J, Chen H, Chen X, et al. Med Sci Monit. Expression of Tumor-Related Macrophages and Cytokines After Surgery of Triple-Negative Breast Cancer Patients and its Implications. 2016; 22:115-20.
4. Yuan ZY, Luo RZ, Peng RJ, et al. High infiltration of tumor-associated macrophages in triple-negative breast cancer is associated with a higher risk of distant metastasis. Onco Targets Ther. 2014;21(7):1475-80.
5 Simoes RV, Serganova IS, Kruchevsky N, et al. Metabolic plasticity of metastatic breast cancer cells: adaptation to changes in the microenvironment. Neoplasia. 2015; 17(8):671-84.
Figures
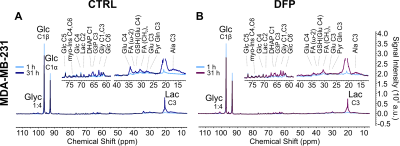
Comparison of representative 13C MR spectra of untreated, control (CTRL, A) and Deferiprone-treated (DFP, B) MDA-MB-231 cells (B). For each treatment condition, the comparison between spectra acquired at the start (1 h; light blue lines; A and B) and the end (31 h; dark blue line, A; prune line; B) of the perfusion experiment is shown. The same vertical scale was used to display all spectra.
Major peak assignments: glycogen (Glyn), glucose (Glc); glutamate (Glu), lactate (Lac), alanine (Ala); dihydroxyacetone phosphate (DHAP); glycerol triphosphate (G3P).
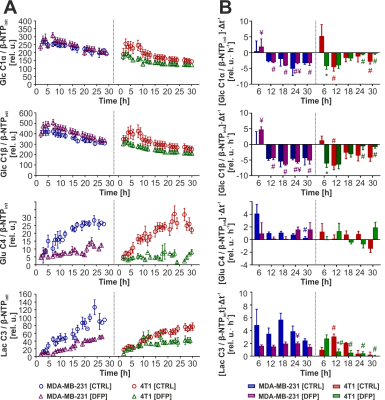
A Time-course effect of Deferiprone (DFP) on cell metabolism of human MDA-MB-231 (left panels) and murine 4T1 (right panel) cells, compared to control (CTRL) cells. B Six-hour average 13C-labeling rates of untreated and DFP-treated MDA-MB-231 (left panels) and 4T1 (right panels) cells.
* Significantly different from CTRL (p < 0.05);
¥ Significantly different from the corresponding value in DFP-treated 4T1 (p < 0.05);
# Significantly different from the value at the 6 h (first) time point for the same cell line and treatment condition (p < 0.05). Colour coded for cell line and condition.
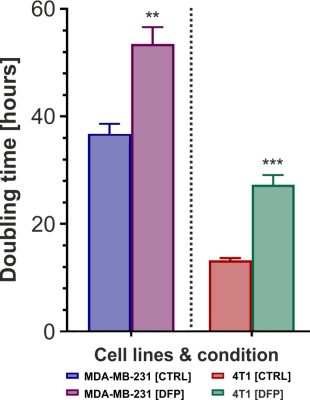
Figure 3. Mean doubling times for untreated (CTRL) and Deferiprone (DFP)- treated MDA-MB-231 (left panel) and 4T1 (right panel) cells.
*** (p < 0.001) and ** (p < 0.01): Significantly different from CTRL (one-tailed, unpaired t-test).
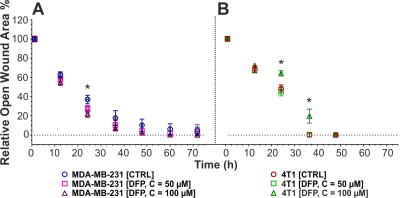
Figure 4. Effect of Deferiprone (DFP) exposure on cell migration of MDA-MB-231 (A) and 4T1 (B) cells. The wound closure is shown as a function of no DFP exposure (CTRL; A, blue circle; B, maroon circle), 50 µM of DFP (A, violet square; B, lime square), and 100 µM of DFP (A, prune triangle; B, green triangle). Data in A and B are averaged over 2-6 independent experiments with 3-6 replicates each and reported as mean ± SD.
* Significantly different from CTRL (p < 0.05, two-way ANOVA followed by Tukey’s post hoc test).
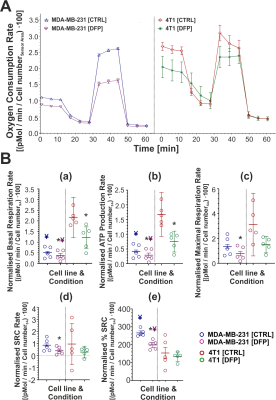
A. Representative time course of oxygen consumption rates in untreated (control, CTRL) or DFP-treated MDA-MB-231 (left graph) and 4T1 (right graph) cells. Values were normalised to the cell number in the sensor area (SA) of each well. B. Summary graphs of five quantitative mitochondrial parameters for MDA-MB-231 (left) and 4T1 cells (right).
* Significantly different from CTRL (p < 0.05);
¥ Significantly different from the corresponding value in DFP-treated 4T1 (p < 0.05);
¥ Significantly different from the corresponding value in CTRL 4T1 (p < 0.05).