4416
Quantification of Electrostatic Molecular Binding Model Using the Water Proton Signal1F.M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States, 2The Russell H. Morgan Department of Radiology, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 3Department of Information Science and Technology, Northwest University, Xi'an, China, 4Key Laboratory for Magnetic Resonance and Multimodality Imaging of Guangdong Province, Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, ShenZhen, China
Synopsis
Saturation transfer MRI has previously been used to probe molecular binding interactions with signal enhancement via the water signal (IMMOBILISE approach). Here, we detailed the relayed nuclear Overhauser effect (rNOE) based mechanisms of this signal enhancement by a four-pool magnetization transfer model, verified the model both using simulations and experimentally (using small charged molecules: arginine, choline, and acetyl-choline). The analytical model can be used to quantify molecular binding affinity, i.e., the dissociation constant (KD). The characterization of the transient binding of small natural substrates paves a pathway towards the detection of receptor-substrate binding in vivo using MRI.
Introduction
Electrostatic interactions play a fundamental role in mediating molecular interactions and are essential in many biochemical processes.1 It was previously shown that transient molecular binding can be imaged using a saturation transfer (ST) approach for enhancing sensitivity,2 the so-called IMMOBILISE (for “IMaging of MOlecular BInding using Ligand Immobilization and Saturation Exchange”) method. More recently, it was shown that this approach can be used to image the binding of small charged molecules via electrostatic interactions to immobile ionic receptors.3Here, the mechanism of water signal enhancement through molecular binding was quantitatively described and verified using simulations and experimentally using select small charged molecules (arginine, choline, and acetylcholine). The derived model was used to quantify binding affinities for these molecules based on the IMMOBILISE signal.
Materials and Methods
TheoryIn the “IMMOBILISE” approach2, the aliphatic protons of free ligands are first selectively labelled using a radio-frequency (RF) pulse, and the signal labelling is transferred to water via several possible binding-mediated pathways (Figure 1). Within free fast-tumbling ligands, intramolecular NOE transfer is very weak. However, upon binding to immobile receptors, the cross-relaxation becomes very efficient (spin diffusion) thus allowing the labelled ligands to couple to water. The following four-pool model was proposed to describe the “IMMOBILISE” process,
HaL k'on↔koff HaLRHe σae↔σea HaLRHe kew↔kwe Hw
where kon and koff are ligand binding on/off rates. At equilibrium, kon[R][L]=koff[LR], where [R], [L] and [LR] are the concentrations of free ligands, free receptor, and bound receptor, respectively. Total receptor concentration [RT] = [LR] + [R]. and σae are σea the effective cross relaxation rates from receptor-bound-ligand aliphatic protons (HaLR) to either exchangeable protons (LRHe) or bound water in the macromolecule and back. kew and kwe are effective saturation transfer rates from LRHe to free water (Hw) and back, corresponding to either the proton or water molecular exchange rates.
The evolution of the z-magnetization for each pool can be described by a set of modified Bloch equations.3 The analytical solution of the IMMOBILISE signal was obtained:
$$$IMMOBILISE={rNOE}_{max}\bullet\frac{\left[L\right]}{\left[L\right]+K_D} $$$ (1)
where rNOEmax is a constant that determines the maximum detected signal, KD ($$$K_D=\frac{k_{off}}{k_{on}}$$$), the dissociation constant.
Sample preparation
Various concentrations (10, 20, 50, 100, and 200 mM) of arginine (Arg), choline (Cho), and acetyl-choline (ACh) were dissolved in phosphate-buffered saline (PBS), pH of 7.2 and mixed with ion-exchange media (Macro-Prep CM Support, with functional groups -COO-). The mixture was pH-adjusted and transferred to the imaging tube for ST measurements.
CEST MRI
Experiments were performed on a Bruker Avance III 17.6 T vertical bore MR scanner at 20°C, using a 4 s CW saturation pulse followed by RARE MRI readout. Z-spectra were acquired within ±6 ppm relative to water in steps of 0.1 ppm, and normalized by the signal intensity without saturation. The B1 was varied from 0.3 to 2.0 μT. Z-spectral components were analyzed using multi-Lorentzian fitting as described previously.4
Results
Using the four-pool Bloch equation modeling, numerical simulations were performed (Figure 2). CEST Z-spectra were shown for binding equilibria with varied free ligand concentration ([L]), binding affinity (KD), and exchange rate (kew), respectively. The simulated IMMOBILISE signal increases with [L] and reaches a plateau when receptors get saturated; is highest with a KD in the mM range; and depends on kew. IMMOBILISE signal is detectable for binding events with slow binding rates ( k'on, within about 101 to 106 s-1), medium affinity (KD, within about 10-7 to 1 M), and above sub-millimolar level of receptor sites ([RT], larger than about 10-4 M). The analytical solution for IMMOBILISE signal (Eq. 1) is also verified by numerical simulations.The results of IMMOBILISE experiments showing the interaction between Arg, Cho, and ACh and negatively charged ion-exchange resin (-COO- functional group) are in Figure 3. The Z-spectra show the detection of Arg aliphatic protons at -0.9, -1.6, and -2.0 ppm, Cho aliphatic protons at -1.6 ppm, and ACh aliphatic protons at -1.6 and -2.6 ppm. ACh has no exchangeable protons, proving one pathway to be intermolecular rNOEs.
The IMMOBILISE signals were measured as a function of free ligand concentration and the curves were fit (Figure 4) using Eq. 1 to estimate KD. The results (Table 1) show that the KD values fitted from three individual peaks of Arg were the same within error (~130 mM). ACh has a higher binding affinity (smaller KD, ~70 mM for the aliphatic proton at -1.6 ppm) with the resin (-COO-) compared to Cho (~160 mM).
Discussion
The quantification of electrostatic molecular binding using IMMOBILISE MRI was demonstrated using simulations and small charged molecules. Simulation results show that molecules with medium affinity (within about 10-7 to 1 M) can provide detectable IMMOBILISE signals, which is comparable to the KD of Arg, Cho, and Ach for the resin binding studied here. Experimentally, we showed that the molecular binding of sub-mM to mM level concentration of receptor sites to molecular targets can be detected by the signal enhancement mechanism, enabling the possibility of in vivo studies of molecular binding using IMMOBILISE MRI.Acknowledgements
No acknowledgement found.References
- Nakamura H. Roles of electrostatic interaction in proteins. Q Rev Biophys. 1996;29:1-90.2.
- Yadav NN, Yang X, Li Y, Li W, Liu G, van Zijl PC. Detection of dynamic substrate binding using MRI. Sci Rep. 2017;7:10138.3.
- Zhou Y, Van Zijl PC, Xu J, Liu X, Yadav N. Detection of ionic bonding using IMMOBILISE MRI and theoretical description of the relayed NOE transfer mechanism. ISMRM; 2021;0243.
- Zhou Y, van Zijl PCM, Xu J, Yadav NN. Mechanism and quantitative assessment of saturation transfer for water-based detection of the aliphatic protons in carbohydrate polymers. Magn Reson Med. 2021;85:1643-1654.
Figures
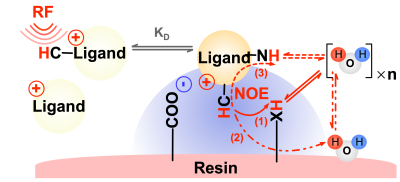
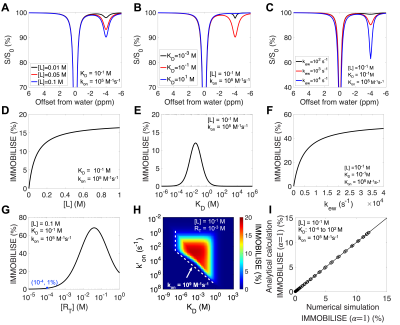
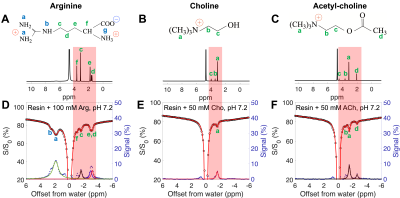
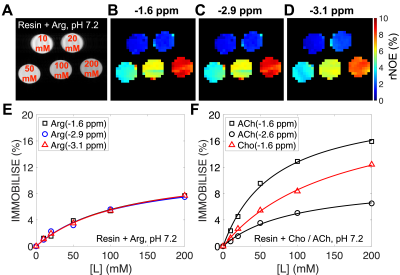
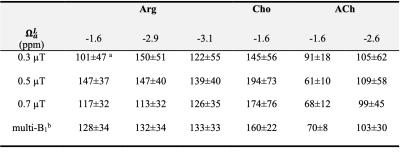
Table 1. Estimated (mM) for L-arginine, Choline, and Acytle-choline binding with ionic resin (-COO-). a Standard deviation (SD) of KD was obtained from 1,000 Monte Carlo simulations of the ligand concentration curve (Eq. 1) with 0.39% random noise, which was estimated from Z-spectra. b Values were obtained by the global fitting of experimental data at all three B1 values simultaneously.