4405
A novel blockface imaging pipeline for robust correlative MR-histology of human brain specimens1Stanford University, Stanford, CA, United States, 2Cornell University, Ithaca, NY, United States, 3Research Centre Jülich, Jülich, Germany
Synopsis
Histology is critical for validating pathology detected in ultra-high-resolution ex-vivo MRI. However, registering histological images with MR is challenging due to 3D deformations between MR and histology, 2D deformations during sectioning, and the fact that histological staining often occurs only in selected slices. Addressing these issues will enable quantification of complex histological features and their relationship with MRI, facilitating the discovery of novel in vivo imaging-based biomarkers of disease. Here, we demonstrate the use of a novel pipeline utilizing ultra-high-resolution specimen MRI, blockface imaging, and a modified tape transfer system for correlative MRI-paraffin-embedded histology of human brain tissue specimens.
Introduction
Determining whether MRI can robustly measure disease-specific micropathology requires precise registration between MRI and histology. However, registering paraffin-embedded histology images with MRI is challenging due to 3D deformations from the paraffin-embedding process in which tissue is dried and embedded in wax. Additional 2D deformations occur during sectioning, mounting, and staining. Furthermore, histological staining is often sparse. Addressing these registration challenges will enable validation of the molecular basis of ex vivo MRI features.We have developed a novel data acquisition and registration pipeline utilizing ultra-high-resolution specimen MRI, including quantitative susceptibility mapping (QSM), Brewster’s angle telecentric blockface imaging to account for distortion during embedding, and a tape-transfer system to minimize the distortion and artifacts resulting from sectioning and slide capture, with automated deformable image registration methods. This pipeline has been optimized and validated on two human brain specimens, one visual cortex (V1), and one hippocampus (HP).
Methods
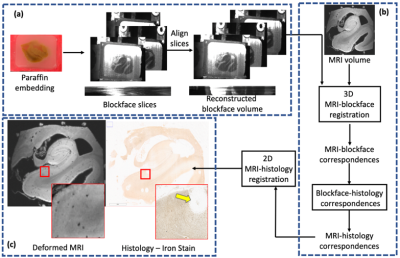
Tissue Preparation:V1 and HP specimens from a patient with Alzheimer’s disease were acquired from the Stanford Alzheimer’s Disease Research Center. For MRI, specimens were immersed in fomblin with a plastic scaffold to minimize motion. Figure 1 shows our pipeline, which consists of acquisition of MRI, blockface, and histology images followed by blockface volume reconstruction, 3D registration of resulting volume and MRI, and 2D pairwise registrations of histology slices and corresponding MRI slices.
Image Acquisition:
Using a Bruker 7T scanner and small-bore millipede coil, ultra-high-resolution 3D-multi-echo gradient echo MRI (0.1mm isotropic, 6 TEs, TR=64ms, Flip angle=22, 3 acquisitions) was acquired. To minimize effects of motion between acquisitions and the difficulty inherent to averaging phase maps across acquisitions, PCA-based denoising across the acquisitions was performed [1], and the first denoised acquisition was used subsequently. We performed a voxel-by-voxel fitting of signal vs. echo time to compute R2* maps, and used the morphology enabled dipole inversion (MEDI) method [2] to compute QSM.
The block was then paraffin-embedded and sectioned at 10-um thickness using a manual microtome. During sectioning, optical blockface images were acquired at the Brewster’s angle (to eliminate the shadow of the tissue under the paraffin) using a telecentric lens (to adjust for barrel and perspective distortion) [3] and polarized filter (to enhance tissue contrast). Sections were captured using either traditional water bath flotation or a novel tape-transfer system tailored to paraffin-embedded sections. Slides were stained for either H&E or iron (DAB-enhanced Perls’ Prussian blue).
Image Processing and Registration:
Blockface volume reconstruction:
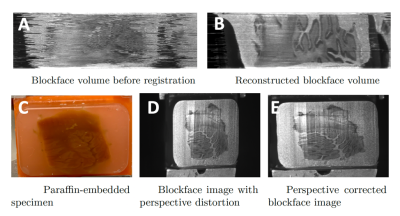
The positions of the camera and microtome can suffer from small perturbations. Therefore, we used a groupwise registration approach, which optimizes image intensity correlations between neighboring slices, to align blockface images before further processing. The telecentric lens does not completely eliminate distortion. To correct for the resultant change in aspect ratio, we adhered a grid with known spacing to the microtome and imaged both face-on and from the Brewster’s angle. The resultant transformation matrix computed between the two images was computed to correct any residual distortion in the blockface images. This procedure is performed once.
Registration of MRI and blockface images:
With the blockface now constituting an intermediary volume, we used an intensity-based affine registration for alignment with MRI, with a mutual information similarity metric.
Registration of MRI to histology:
Given the 3D correspondence between MRI and blockface images, and that each blockface image corresponds to a histological slice, the final step is a slice-wise registration between the histological section and corresponding MRI slice. After extracting the appropriate MRI slice for an unaligned histology slice, we perform a two-stage (linear, then nonlinear) intensity-based registration using a mutual information metric.
Results
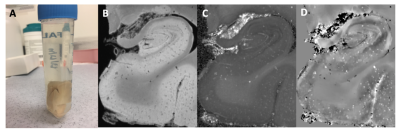
MRI:Processing the mGRE sequence produces high-resolution denoised structural and QSM imaging (Fig. 2).
Blockface Volume Reconstruction:
Figure 3 illustrates the significant benefits of the groupwise registration procedure in the blockface volume reconstruction. The calibration step effectively reduces residual distortion (Fig. 3).
Tape-Transfer Validation:
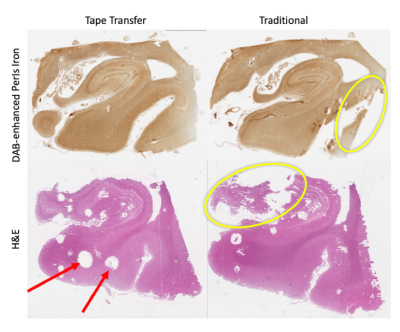
Our novel tape transfer process provides marked improvement in the quality of resultant histology with more complete coverage of the tissue with fewer tears, folds, and artifacts, though tape transfer slides are not completely distortion-free (Fig. 4). Separately, we verified that tape-transfer does not affect staining.
MR-Histology Registration:
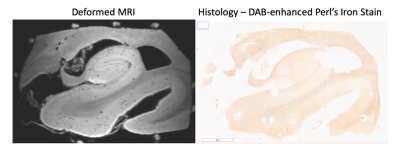
Example slices for both histological stains were aligned to the MRI to validate the registration for stains with differing contrasts and signal density. Figure 5 presents example registration between MRI and iron stain for the HP, with clear visual improvement from the unaligned slice.
Discussion
We implemented an integrated pipeline for correlative ex vivo MRI-histology. By utilizing blockface imaging leveraging specific properties of polarized optics to generate enhanced contrast in the blockface images, we have improved the slice-to-slice correspondence between MRI and individual histology images. Through optimization of a new tape-transfer method for capturing sections, we have drastically reduced distortion and artifacts. Automated deformable registration methods facilitate higher-throughput processing than prior techniques.Conclusion
This pipeline may enable validation of MRI features and development of novel biomarkers. We next plan to utilize this pipeline for robust coregistration of focal subicular hypointensities found on MRI with histological stains for iron, microglia, amyloid, and tau, to validate MRI-based measures of iron-containing microglia as a biomarker in Alzheimer’s disease.Acknowledgements
The authors would like to acknowledge the continued support of the Stanford Alzheimer’s Disease Research Center (ADRC) for this research, as well as the NIH, Doris Duke Charitable Foundation and the Dana Foundation for their funding.References
[1] Veraart, J., Novikov, D. S., Christiaens, D., Ades-Aron, B., Sijbers, J., & Fieremans, E. (2016). Denoising of diffusion MRI using random matrix theory. Neuroimage, 142, 394-406.
[2] Liu, J., Liu, T., de Rochefort, L., Ledoux, J., Khalidov, I., Chen, W., ... & Wang, Y. (2012). Morphology enabled dipole inversion for quantitative susceptibility mapping using structural consistency between the magnitude image and the susceptibility map. Neuroimage, 59(3), 2560-2568.
[3] Shojaii, R., Bacopulos, S., Yang, W., Karavardanyan, T., Spyropoulos, D., Raouf, A., ... & Seth, A. (2014). Reconstruction of 3-dimensional histology volume and its application to study mouse mammary glands. Journal of visualized experiments: JoVE, (89).
Figures