4401
ACID - an open-source, bids compatible software for brain and spinal cord dMRI: preprocessing, DTI/DKI, biophysical modelling1Department of Systems Neuroscience, University Medical Center Hamburg-Eppendorf, Hamburg, Germany, 2Balgrist University Hospital, University of Zurich, Zürich, Switzerland, 3Department of Mathematics, Emory University, Atlanta, GA, United States, 4Weierstrass Institute for Applied Analysis and Stochastics, Berlin, Germany, 5Department of Neurophysics, Max Planck Institute for Human Cognitive and Brain Sciences, Leipzig, Germany
Synopsis
The ACID-Toolbox is an open-source toolbox for brain and spinal-cord diffusion data. It enables the preprocessing and creation of DTI/DKI, and biophysical parameter-maps in one single pipeline. Preprocessing covers correction for eddy-currents and motion-artefacts, susceptibility distortions and adaptive denoising. For model-fitting, the toolbox offers several algorithms to estimate the diffusion or the kurtosis tensors as well as estimation of biophysical parameters. ACID is integrated into the batch system of the Statistical-Parametric-Mapping (SPM12) software for the analysis of neuroimaging data and thus benefits from associated spatial and statistical processing modules. Additionally, ACID is BIDS compatible but also applicable to non-BIDS-conform data.
Introduction
Diffusion MRI (dMRI) has become an important tool for neuroscientific and clinical research due to its sensitivity to the microstructure of brain and spinal cord tissue $$$^{1,2}$$$. dMRI is highly sensitive to subject motion, physiological noise, and suffers from the typical artifacts of the echo-planar imaging (EPI) sequence (ghosting, distortion, blurring, etc.) $$$^{3,4,5}$$$. For the brain, several toolboxes have been developed to address these artefacts and process dMRI data, but the majority of these tools are not applicable or optimized for the spinal cord. The Artefact Correction in Diffusion MRI (ACID) toolbox is a MATLAB-based open-source academic software toolkit that provides retrospective artifact correction methods for both brain and spinal cord dMRI data. ACID is integrated into the batch system of the Statistical Parametric Mapping (SPM12) software for the analysis of neuroimaging data and thus benefits from associated spatial and statistical processing modules. Besides artifact correction, ACID provides the tools for fitting the diffusion and kurtosis tensor, estimating biophysical parameters such as axon-water fraction or fiber dispersion, and dedicated postprocessing and analysis solutions.Methods
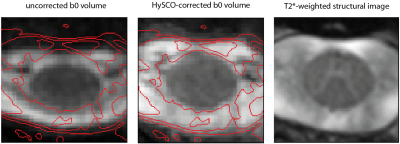
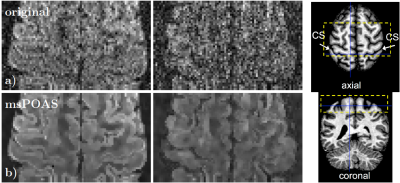
The ACID Toolbox is available under http://www.diffusiontools.com/ along with a tutorial.Pre-processing: ACID contains three retrospective artifact correction tools. (a) Eddy Current and Motion Correction (ECMOCO) is designed for correcting affine displacements due to eddy current artifacts and subject motion (Fig. 1) $$$^3$$$. (b) Hyperelastic Susceptibility Artifact Correction (HySCO) is a tool for correcting susceptibility artifacts based on a reversed gradient-based acquisition scheme (Fig. 2) $$$^6$$$. (c) Multi-shell position-orientation adaptive smoothing (msPOAS) is an adaptive denoising method which is compatible with all diffusion models and therefore universally applicable (Fig. 3) $$$^5$$$.
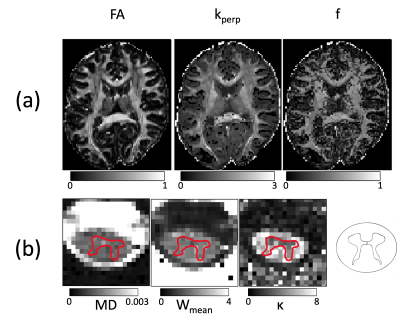
Tensor fitting: For model fitting, ACID offers several algorithms to estimate the diffusion (DTI) or the kurtosis tensor (DKI) parameters (Fig. 4a) including ordinary and weighted least squares, robust tensor fitting, nonlinear model fitting (NNL), and Rician bias corrected parameter estimation.
Biophysical models: ACID contains two biophysical models that estimate biophysical parameters associated with the “standard tissue model” using either DTI $$$^8$$$ or DKI $$$^9$$$ parameters. These include the axon-water fraction (f) and fiber dispersion ($$$\kappa$$$) (Fig. 4b), as well as intra- and extra-axonal diffusivities ($$$D_a$$$, $$$D_{e\perp}$$$, and $$$D_{e\parallel}$$$).
Post-processing: For image post-processing, ACID has its own functionalities but also relies on SPM tools, e.g., for non-linear spatial alignment. Two modules dedicated to the spinal cord are of particular interest here: (a) a semi-automated tool to refine spatial registration at group level and (b) reliability masking to automatically identify “unreliable" voxels, i.e., voxels affected by irreversible motion artifacts. Subsequently, the identified voxels can be excluded from the analysis in the form of subject-specific “reliability masks".
Pipelines: The presented tools can be combined into linear pipelines, where subsequent steps rely on the output of previous steps and the intermediated results can be transferred to the next module via flexible dependencies. For example: raw dMRI first undergo eddy currents and motion artifact correction, followed by adaptive smoothing (msPOAS), susceptibility artifact correction (HySCO), and model fitting.
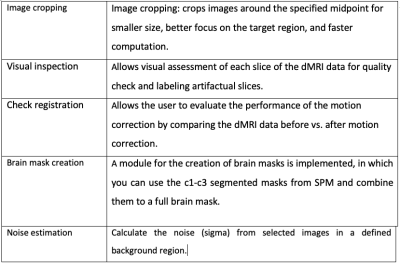
ACID utilities: The ACID Toolbox features several utility tools for quality check and improved usability. The most important utilities are displayed in Table 1.
BIDS: ACID supports the Brain Imaging Data Structure (BIDS) format, meaning that BIDS-compliant output is generated for BIDS-compliant input. This includes the creation of separate bval and bvec files, as well as a json file in which the measurement parameters and previous processing steps are documented. For non-BIDS data, the file naming and folder structure is treated as quasi-BIDS. The output is stored in a derivatives folder containing the results (e.g., ECMOCO-Run_”Number of Run”).
Results and Discussion
We demonstrated ACID on an example brain and spinal cord dMRI dataset acquired in a healthy volunteer on a 3T Siemens Prisma-fit using a single-shot echo planar imaging sequence. Following sequence parameters were used for brain: 151 diffusion-sensitizing gradient directions and b-values of 0, 500, 1250 and 2500 $$$\frac{s}{mm^2}$$$. FOV=240x230x154 $$$mm^3$$$, resolution=1.6 $$$mm^3$$$ isotropic, TE/TR=73ms/5300 ms. Following sequence parameters were used for spinal cord: 101 diffusion-sensitizing gradient directions and b-values of 0, 500, 1000 and 1500 $$$\frac{s}{mm^2}$$$. FOV=128x36x70 $$$mm^3$$$, resolution=1x1x5 $$$mm^3$$$, TE/TR=73ms/260ms. dMRI data underwent ECMOCO, HySCO, POAS correction, and were fitted a nonlinear standard DKI model with Rician bias correction to obtain the 21 standard DKI parameters. Finally, biophysical parameters were estimated. The output of HySCO and msPOAS are displayed in Fig. 2 and Fig. 3, respectively. Both modules results in visible improvements of the images.This illustrates the usability of the toolbox for pre-processing as well as the calculation of DTI, DKI and biophysical parameters in one pipeline. The BIDS compatibility unifies the structure of the outputs, while the compatibility to tools which don’t use the BIDS standard have to be ensured.Conclusion
ACID is an easy-to-use open-source add-on to SPM12 which provides an optimized framework to process and analyze brain and spinal cord dMRI data. The toolbox has been developed as a response to the increasing number of studies applying dMRI in the spinal cord, and offers comprehensive tools for artifact correction, model fitting, quality assessment, and analysis. ACID makes it possible to preprocess and create multiparameter maps in one single pipeline, which unifies and simplify the analysis of several similar data sets.Acknowledgements
This work was supported by the German Research Foundation (DFG Priority Program 2041 "Computational Connectomics”, [MO 2397/5-1,2], by theEmmy Noether Stipend: MO 2397/4-1) and by the BMBF (01EW1711A and B) in the framework of ERA-NET NEURON.References
1. Mohammadi, S., Tabelow, K., Ruthotto, L., Feiweier, T., Polzehl, J., and Weiskopf, N.. High-resolution diffusion kurtosis imaging at 3T enabled by advanced post-processing. Front. Neurosci. (2015b) 8:427.
2. David, Gergely, Patrick Freund, und Siawoosh Mohammadi. „The Efficiency of Retrospective Artifact Correction Methods in Improving the Statistical Power of Between-Group Differences in Spinal Cord DTI“. NeuroImage 158 (2017):296–307.
3. Mohammadi S, Moller HE, Kugel H, Muller DK, Deppe M Correcting eddy current and motion effects by affine whole-brain registrations: evaluation of three-dimensional distortions and comparison with slicewise correction. Magn Reson Med 64: (2010):1047-1056.
4. Ruthotto, L, Kugel, H, Olesch, J, Fischer, B, Modersitzki, J, Burger, M, and Wolters, C H. Diffeomorphic Susceptibility Artefact Correction of Diffusion-Weighted Magnetic Resonance Images. Physics in Medicine and Biology, 57(18), (2012):5715-5731.
5. Becker, S.M.A., K. Tabelow, S. Mohammadi, N. Weiskopf, und J. Polzehl. „Adaptive Smoothing of Multi-Shell Diffusion Weighted Magnetic Resonance Data by MsPOAS“. NeuroImage 95 (2014): 90–105.
6. Ruthotto, L., Mohammadi, S., Heck, C., Modersitzki, J., and Weiskopf, N. . “Hyperelastic susceptibility artifact correction of DTI in SPM,” in Bildverarbeitung Für Die Medizin 2013, eds H. P. Meinzer, T. Deserno, H. Handels, and T. Tolxdorff (Heidelberg: Springer), (2013):344–349.
7. Macdonald, J., and Ruthotto, L. . Improved susceptibility artifact correction of echo-planar MRI using the alternating direction method of multipliers. J. Math. Imaging Vis. 60, (2018):268–282.
8. Edwards, Luke J., Kerrin J. Pine, Isabel Ellerbrock, Nikolaus Weiskopf, und Siawoosh Mohammadi. „NODDI-DTI: Estimating Neurite Orientation and Dispersion Parameters from a Diffusion Tensor in Healthy White Matter“. Frontiers in Neuroscience 11 (2017): 720.
9. Jespersen, Sune Nørhøj, Jonas Lynge Olesen, Brian Hansen, und Noam Shemesh. „Diffusion Time Dependence of Microstructural Parameters in Fixed Spinal Cord“. NeuroImage 182 (2018): 329–42.
Figures