4398
Linear+: An optimized sequence reordering for robust scout accelerated retrospective motion estimation and correction1Siemens Healthcare GmbH, Erlangen, Germany, 2A. A. Martinos Center for Biomedical Imaging, Charlestown, MA, United States, 3Siemens Medical Solutions, Malvern, PA, United States, 4Department of Radiology, Massachusetts General Hospital, Boston, MA, United States
Synopsis
Distributed sequence reorderings for 3D multi-shot acquisitions ensure overlap with the central k-space in every shot. This improves the stability of navigator-free retrospective motion estimation but often reduces the robustness of the image reconstruction. In this work, we have developed and optimized a novel sampling strategy (Linear+) that extends the standard sequential sequence ordering with a small number of additional calibration samples acquired near the k-space center. In simulation and in vivo, Linear+ enabled accurate motion parameter estimation and correction across multiple motion patterns while providing improved image homogeneity and spatial resolution compared to a distributed reordering.
Introduction
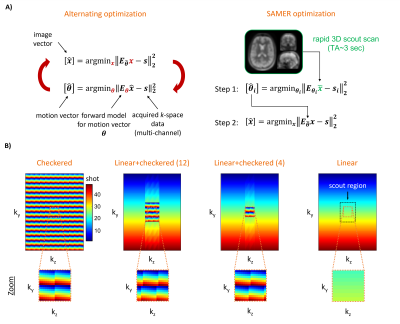
Navigator-free retrospective motion correction techniques1–4 often solve a coupled optimization problem (Fig. 1A) where the data-consistency error of a SENSE+motion forward model is minimized with respect to the unknown motion parameters and an image estimate “jointly”. This poses a computationally demanding non-convex inverse problem with several hundred temporal motion parameters and millions of imaging voxels. The recently proposed SAMER technique5 leverages an ultra-fast, low-resolution scout scan as an image prior. This strategy decouples motion estimation from the image reconstruction and avoids the computationally demanding repeated updates of an image estimate (Fig. 1A).Optimized encoding reorderings for rectilinear 3D multi-shot acquisitions, such as checkered6 or linear+checkered5 sampling have been shown to improve the stability of motion estimation by ensuring that each shot has distributed overlap with the central k-space region (Fig. 1B). Unfortunately, while improving the motion parameter estimation, it adversely effects the image pixel estimation. To overcome these limitations, we have developed a novel sampling strategy that retains the stability of SAMER motion estimation while allowing for robust image reconstructions.
Methods
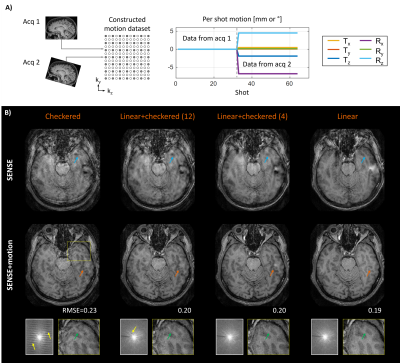
Sequence reordering and effect on SENSE+motion reconstruction:We first analyzed how the sequence reordering affects the image quality in SENSE+motion reconstructions. For this, two motion-free MPRAGE scans were acquired at different head poses and the acquired k-space data was combined to generate artifical motion corrupted datasets. This was done with checkered, linear+checkered (12), linear+checkered (4) and standard linear sampling (Fig. 1B). For each dataset, SENSE and SENSE+motion reconstructions were performed using ground truth motion parameters obtained from a direct registration of the motion-free imaging volumes. All scans were acquired on a 3T system (MAGNETOM Skyra, Siemens Healthcare, Erlangen, Germany) using a 20-channel head coil.
Optimization of Linear+ sequence reordering:
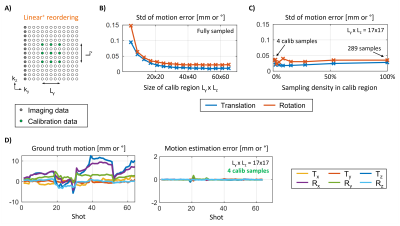
As can be seen in Fig. 2, linear sampling is most favorable for SENSE+motion reconstructions. However, as previously demonstrated the lack of k-space center information in every shot (Fig. 1B) impedes accurate motion estimation5. To bridge the image quality gap between linear+checkered (4) and linear we have developed a novel reordering, Linear+, and reconstruction strategy. Here, imaging data with linear reordering is combined with a fixed set of calibration samples acquired in each shot near the center of k-space (Fig. 3A). Through simulation, we investigated the minimal extent and sampling density required to achieve accurate motion estimation. Note, the calibration samples are only used for motion estimation and discarded during the SENSE+motion reconstruction to avoid reconstruction instabilities (Fig. 2B).
In vivo motion experiments:
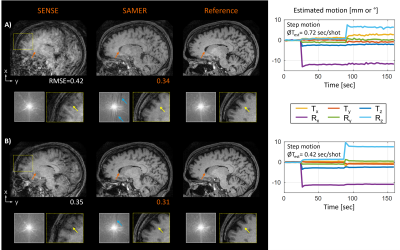
Figure 3 suggests our optimized Linear+ reordering requires only four additional calibration samples per shot. To accommodate for this additional 2% of data sampling, we reduced the nominal spatial resolution along the partition direction by the same percentage as this avoids any changes to the sequence timing while having minimal effect on image sharpness. Moreover, the small extent of the calibration region in k-space (L=17x17) allowed us to further reduce the spatial resolution of the 3D MPRAGE scout scan5 from 1x4x4mm³ to 1x8x8 mm³ allowing its acquisition in just a single TR=2.5 sec of additional scan time. On one healthy volunteer, we directly compared the image quality of Linear+ reordering to standard linear+checkered sampling using instructed step motion and further investigated the performance of the Linear+ approach across three clinically representative motion patterns.
Results
The SENSE+motion reconstruction of checkered sampling showed inhomogeneous contrast and loss of spatial resolution due to extensive gaps in k-space (Fig. 2B). These issues were reduced by linear+checkered sampling where the artifact level improved as the size of the checkered region decreased. However, even when considering linear+checkered (4) the image quality and RMSE were below that of linear reordering.The motion estimation error in Linear+ reordering rises sharply as the calibration extent is restricted (Fig. 3B). In contrast, reducing the sampling density within the calibration region (fixed calibration size) does not significantly affect the motion estimation accuracy (Fig. 3C). Using only four calibration samples Linear+ reordering enabled accurate motion parameter estimation with negligible error (Fig. 3D).
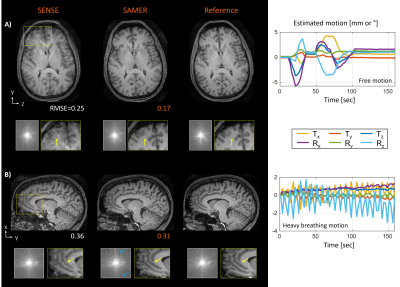
The in vivo acquisitions with Linear+ reordering showed more homogenous contrast and better spatial resolution than linear+checkered sampling (Fig. 4). Moreover, the average motion estimation time decreased (linear+checkered: 0.72 sec/shot, Linear+: 0.42 sec/shot) due to the lower spatial resolution of the scout. In Fig. 5, robust image reconstruction using Linear+ reordering is demonstrated across multiple clinically representative motion trajectories.
Discussion
Limiting checkered sampling to a small region of k-space, as in linear+checkered (4), reduced gaps in k-space and loss of spatial resolution but it did not achieve the image quality of linear sampling. The remaining image quality gap was caused by orientation-dependent phase differences across the motion states7, which checkered sampling is highly sensitive to. Our Linear+ reordering avoids checkered sampling and the difficult task of estimating phase variations across motion states. Our optimized reordering relies on linear sampling and a small number of calibration samples that are discarded during the image reconstruction (2% resolution loss). In our in vivo evaluations, this strategy improved the spatial resolution and image homogeneity in a direct comparison against linear+checkered sampling and achieved good motion correction performance across a variety of representative motion trajectories.Acknowledgements
This work was supported in part by NIH research grants: 1P41EB030006-01, 5U01EB025121-03References
1. Haskell MW, Cauley SF, Wald LL. TArgeted Motion Estimation and Reduction (TAMER): Data consistency based motion mitigation for mri using a reduced model joint optimization. IEEE Trans Med Imaging. 2018;37(5):1253-1265. doi:10.1109/TMI.2018.2791482
2. Loktyushin A, Nickisch H, Pohmann R, Schölkopf B. Blind multirigid retrospective motion correction of MR images. Magn Reson Med. 2015;73(4):1457-1468. doi:10.1002/mrm.25266
3. Cordero-Grande L, Teixeira RPAG, Hughes EJ, Hutter J, Price AN, Hajnal J V. Sensitivity Encoding for Aligned Multishot Magnetic Resonance Reconstruction. IEEE Trans Comput Imaging. 2016;2(3):266-280. doi:10.1109/tci.2016.2557069
4. Cordero-Grande L, Hughes EJ, Hutter J, Price AN, Hajnal J V. Three-dimensional motion corrected sensitivity encoding reconstruction for multi-shot multi-slice MRI: Application to neonatal brain imaging. Magn Reson Med. 2018;79(3):1365-1376. doi:10.1002/mrm.26796
5. Polak D, Splitthoff DN, Clifford B, et al. Scout accelerated motion estimation and reduction (SAMER). Magn Reson Med. n/a(n/a). doi:https://doi.org/10.1002/mrm.28971
6. Cordero-Grande L, Ferrazzi G, Teixeira RPAG, O’Muircheartaigh J, Price AN, Hajnal J V. Motion-corrected MRI with DISORDER: Distributed and incoherent sample orders for reconstruction deblurring using encoding redundancy. Magn Reson Med. 2020;84(2):713-726. doi:10.1002/mrm.28157
7. Brackenier Y, Cordero-Grande L, Tomi-Tricot R, et al. Data-driven motion-corrected brain MRI incorporating pose dependent B0 fields. In: Proceedings of ISMRM 29th Annual Meeting. ; 2021.
Figures