4396
Retrospective Motion Artifacts Suppression by Simulated radial K-space (R-MASSK) for 3D view-sharing sequence acquired abdominal 4D-MRI1Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, Hong Kong, 2Department of Biomedical Engineering, The Chinese University of Hong Kong, Hong Kong, Hong Kong
Synopsis
3D view-sharing sequence has been demonstrated its great promise for abdominal 4D-MRI due to its high temporal resolution and availability in clinical scanners. However, the sub-optimal use of these sequence in free-breathing situation deteriorates the image quality by ghost artifacts induced by respiration. We propose a novel technique, Retrospective Motion Artifacts Suppression by Simulated radial K-space (R-MASSK), to suppress motion artifacts and to increase image quality of the 4D-MRI acquired by the 3D view-sharing sequences, TRICKS and TWIST, through simulated radial k-space resampling.
Purpose
4D-MRI is an important technique for respiratory motion management in radiotherapy. The clinical adaption of 4D-MRI sequences is generally hindered by the low image quality, motion artifacts, and clinical availability. This study aims to propose a novel technique, Retrospective Motion Artifacts Suppression by Simulated radial K-space (R-MASSK), that can suppress motion artifacts and enhance image quality of clinically available 3D view-sharing 4D-MRI.Methods
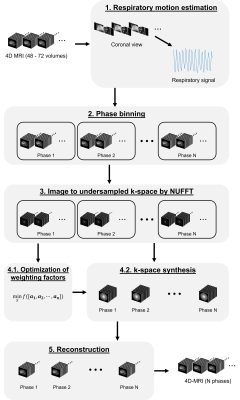
The overview of the method is summarized as a schematic diagram in Fig. 1. In brief, the time-resolved 4D-MRI images were first sorted into 6 or 8 phase based on the amplitude of the surrogated signal. The sorted images were then converted to undersampled golden-angle radial k-space. A radial k-space, with motion averaging effect and suppressed motion artifacts, was synthesized for each respiratory phase based on the optimization of weighting factors by iterative sequential quadratic programming (SQP) method. 1 After an iterative image reconstruction, a respiratory phase-resolved 4D-MRI with suppressed motion artifacts, and improved image quality was obtained.Patient study: The R-MASSK technique was tested on clinically acquired 4D-MRI images of 20 liver cancer patients using 3D view-sharing sequences – TRICKS or TWIST, on a 1.5T or 3.0T scanners (10 patients for each scanner), respectively.
Quantitative evaluation: The R-MASSK 4D-MRI, result-driven (RD) sorted 4D-MRI, 2 RD sorted 4D-MRI followed by the sequential wavelet denoising (RD+D), and the original 4D-MRI were then evaluated by signal-to-noise ratio (SNR), hepatic portal vein-to-liver parenchyma contrast-to-noise ratio (CNR), and gradient energy (GE) percentage decrease.
Results
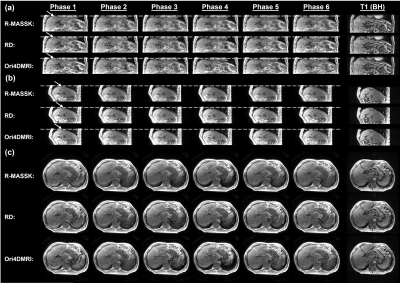
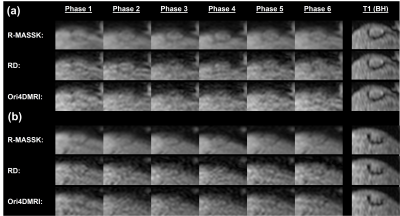
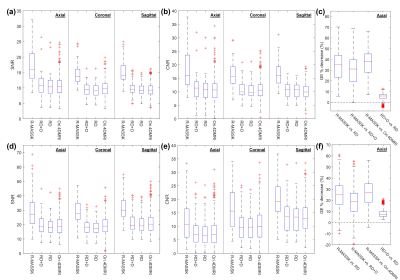
Fig.2 shows the R-MASSK 4D-MRI of a representative patient scanned by TRICKS sequence on the 1.5T scanner. The 4D-MRI sorted by RD strategy and the original 4D-MRI are also presented in Fig.2 as comparison. The proposed R-MASSK technique shows significant improvement on motion artifacts and overall image quality in all three planes (coronal, sagittal, and axial), and all respiratory phases, as compared to the RD-sorted 4D-MRI and the original 4D-MRI. For instance, a tumor located on hepatic dome can be clearly visualized on the R-MASSK 4D-MRI, but poorly identified on the RD 4D-MRI and on the original 4D-MRI, as shown in the magnified images in Fig.3.Quantitative measurements of R-MASSK 4D-MRI, RD 4D-MRI, and original 4D-MRI of 20 patients (10 for each cohort) are presented as boxplots in Fig. 4. R-MASSK 4D-MRI shows higher SNR and CNR in all three planes as compared to the RD+D, RD, original 4D-MRI. For cohort of patients scanned on the 1.5T scanner, the average magnitude of SNR (CNR) for R-MASSK, RD+D, RD 4D-MRI, and original 4D-MRI over all three planes are 15.7 ± 4.50 (17.0 ± 5.62), 10.1 ± 2.91 (10.93 ± 3.69), 9.77 ± 2.75 (10.56 ± 3.56), and 10.07 ± 2.77 (10.59 ± 3.72), respectively. For cohort of patients scanned on the 3.0T scanner, the average magnitude of SNR (CNR) for R-MASSK, RD+D, RD 4D-MRI, and original 4D-MRI over all three planes are 29.6 ± 9.70 (16.2 ± 8.13), 19.9 ± 6.73 (10.7 ± 6.12), 19.1 ± 6.29 (10.3 ± 5.85), and 20.0 ± 6.83 (10.36 ± 6.10), respectively. While GE percentage decrease can effectively quantify the ghost artifacts on the 4D-MRI images resulted from the physiological motion. 3 R-MASSK 4D-MRI also shows significant reduction in GE as compared to the RD+D, RD, original 4D-MRI. For cohort of patients scanned on the 1.5T scanner (3.0T scanner), the average magnitude of GE percentage decrease for R-MASSK 4D-MRI with respect to RD, RD+D, and original 4D-MRI are 34.0 % ± 13.21 % (25.12 % ± 11.65 %), 30.19 % ± 13.34 % (18.45 % ± 11.73 %), and 36.42 % ± 12.37 % (26.57 % ± 11.69 %), respectively.
Discussion
3D view-sharing sequence, such as TRICKS and TWIST, which originally was designed for dynamic contrast enhanced MRI or MRA, has been demonstrated its potential application in abdominal 4D-MRI due to its ultra-fast acquisition speed (< 1 s/volume). 4,5 However, the low acquisition rate of peripheral k-space in these sequences leads to a great vulnerability to respiratory motion, and hence, a decrease in image quality. Therefore, we propose a novel technique that particularly aims to mitigate the motion artifacts of the TRICKS/TWIST acquired 4D-MRI. Moreover, conventional 4D-MRI binning techniques tend to discard redundant frames in each phase bins, resulting in a decrease of the overall efficiency of the scanning procedure. 2,6,7 The proposed R-MASSK technique fully explores the information of the redundant frames by means of retrospective conversion and resampling of radial k-space. Through the optimization of resampling factors by SQP method, the reconstructed 4D-MRI associated with the simulated radial k-space manifests minimal motion artifacts and noise level. 3Conclusion
Both qualitative and quantitative results suggest that R-MASSK technique is capable of suppressing motion artifacts and improving image quality in both axial, coronal, and sagittal planes of the 4D-MRI acquired by 3D view-sharing sequence.Acknowledgements
No acknowledgement found.References
1. Schittkowski K. NLPQL: A fortran subroutine solving constrained nonlinear programming problems. Ann Oper Res. 1986;5(2):485-500.
2. Liu Y, Yin F-F, Czito BG, Bashir MR, Cai J. T2-weighted four dimensional magnetic resonance imaging with result-driven phase sorting. Med Phys. 2015;42(8):4460-4471.
3. Madore B, Henkelman RM. A new way of averaging with applications to MRI. Med Phys. 1996;23(1):109-113.
4. Plathow C, Schoebinger M, Herth F, Tuengerthal S, Meinzer H-P, Kauczor H-U. Estimation of Pulmonary Motion in Healthy Subjects and Patients with Intrathoracic Tumors Using 3D-Dynamic MRI: Initial Results. Korean J Radiol. 2009;10(6):559.
5. Yuan J, Wong OL, Zhou Y, Chueng KY, Yu SK. A fast volumetric 4D-MRI with sub-second frame rate for abdominal motion monitoring and characterization in MRI-guided radiotherapy. Quant Imaging Med Surg. 2019;9(7):1303-1314.
6. Yang Z, Ren L, Yin F-F, Liang X, Cai J. Motion robust 4D-MRI sorting based on anatomic feature matching: A digital phantom simulation study. Radiation Medicine and Protection. 2020;1(1):41-47.
7. Tryggestad E, Flammang A, Han-Oh S, et al. Respiration-based sorting of dynamic MRI to derive representative 4D-MRI for radiotherapy planning. Med Phys. 2013;40(5):051909.
Figures