4384
A user interface for brain cell analysis of a petascale database in cerebral cortex and its application to numerical simulations of diffusion1Northeastern University, Boston, MA, United States, 2Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, MA, United States, 3Department of Radiology, Harvard Medical School, Boston, MA, United States
Synopsis
The 1.4 petabyte electron microscopy volume with fully segmented cells in the human cerebral cortex is publicly available, serving as a valuable resource of building numerical phantoms of MRI. However, even few neurite interruptions in cell segmentations can lead to substantial biases in numerical simulations of diffusion inside cells. Here we propose a user interface to automatically correct most of the neurite interruptions and manually edit the corrections. Combing the proposed user interface with Trees toolbox and SpinDoctor for quantitative cell analysis and diffusion simulations, we provide a convenient platform to validate biophysical modeling of MRI evaluating tissue microstructure.
Introduction
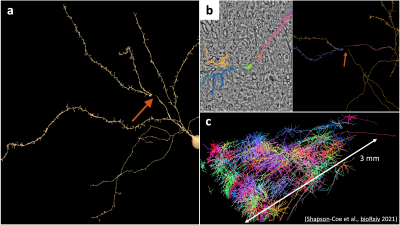
Numerical phantom is a gold standard to validate biophysical modeling of MRI evaluating tissue microstructure [1]. Recently, a 1.4 petabyte electron microscopy (EM) volume with fully segmented cells and cell parts in the human temporal lobe of cerebral cortex is published and publicly available [2], serving as a valuable resource of building numerical phantom in the brain white matter [3] and gray matter (GM). The quality of EM data and cell segmentations is satisfying for the histological analysis. However, even few interruptions in segmented cell shapes (e.g., breaking neurites) can lead to significant biases in numerical simulations of diffusion inside cells (Fig. 1). Here, we propose a user interface with tools to (i) automatically correct most of the neurite interruptions in segmented cells, and manually proofread/edit these corrections, (ii) quantitatively analyze and report the cell features using Trees Toolbox [4], and (iii) perform numerical simulations of diffusion in realistic cell shapes using the neuron module and finite-element method in SpinDoctor [5-6]. With the proposed user interface, we can quickly correct and analyze cell segmentations in the petascle database, and perform diffusion simulations to validate dMRI models, effectively bridging the gap of three orders of magnitude in spatial scales of MRI voxels and tissue microstructure.Methods
Ex vivo EM and cell segmentation in human brainA fresh surgical cerebral cortex sample from the anterior temporal lobe (45yo, F) was quickly preserved, stained, embedded, and sectioned [2]. More than 5000 sequential 30 nm thick sections were collected using an ATUM, and imaged in a multi beam scanning EM. Each section is about 4×4 mm2 in area with 4×4 nm2 pixel size. 196 million tiles of data containing cortex were stitched, aligned, and segmented, yielding a 1.4 petabyte cortex volume. Four proofreaders further completed segmentations of 104 randomly selected neurons for the comparison with un-proofread cells. More details are in ref. [2]. We will demonstrate our cell fixation pipeline in these proofread cells.
User Interface
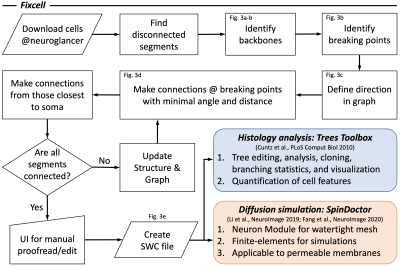
In this petascale EM volume, neurite interruptions in cell segmentations are observed. To fix the cell interruptions, we develop a Matlab-based user interface (UI) for the cell fixation (Fixcell), and further combine the proposed UI with Trees toolbox and SpinDoctor (both in Matlab) for histological analysis and numerical simulations of diffusion (Figs. 2-4):
i. Automatic correction and manual proofreading of neurite interruptions
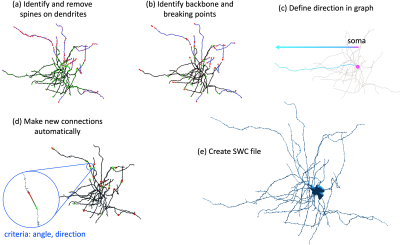
- After downloading the skeleton (nodes, edges, and local radii) of proofread cell in ref. [2] from Neuroglancer, an undirected graph is constructed with its nodes and edges. Each disconnected segment (subgraph) is identified in the graph, and the largest subgraph is considered as the main subgraph to start with the cell fixation.
- To identify the backbone, the spines of dendrites are identified and removed (Fig. 3a). The end nodes of the backbone of each subgraph are considered as the candidate breaking points (Fig. 3b, 4a), where the neurite interruptions happen and need to be fixed.
- To avoid false connections, the edge direction from the soma to the neurite end is defined in the main subgraph (Fig. 3c). From the candidate breaking points of the main subgraph, the new connection is automatically made by choosing the one with the smallest distance between breaking points and the smallest angles between two ends (Fig. 3d, 4b). The connections that are closest to soma is prioritized.
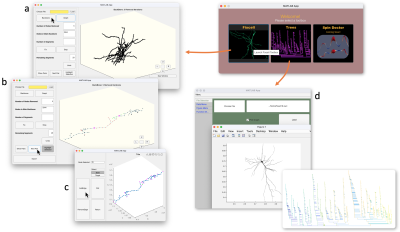
- The main subgraph keeps growing by making new connections until all the segments are connected. Then an UI is provided for the manual proofreading and connection editing (Fig. 4c). Finally, a fully connected cell skeleton is saved an SWC file (Fig. 3e), a commonly used format for quantitative analysis of cells and numerical simulations.
ii. Quantitative analysis of cell features
Trees toolbox [4] offers variety of tools for tree editing, analysis, cloning, branching statistics, and visualization, and has been widely used for quantification of cell features [7] and generative model of cell design [8].
iii. Numerical simulations of diffusion in cells
SpinDoctor [5-6] has the Neuron module to translate the cell skeleton in SWC file into a watertight 3D mesh for finite-element diffusion simulations. As the Laplace eigenfunctions are calculated for a given cell shape and a membrane permeability value, the diffusion signals of any protocols can be quickly simulated. This is particularly suitable for the optimization of diffusion MRI sequence protocols in a timely manner.
Results
The cell fixation pipeline is demonstrated in the cell skeleton of a proofread cell in ref. [2]. In this exemplified cell (Fig. 3), all of the neurite interruptions are successfully corrected with automatic pipeline. In other proofread cells, with appropriate parameter setting, only 2-3 neurite interruptions need to be manually proofread and corrected, substantially reducing the load of proofreading and cell editing. The calculation time is 2-10 mins per cell on a home computer based on the cell size.Discussion and conclusions
The proposed pipeline (Fixcell) automatically corrects most of the neurite interruptions in cell skeletons with few tunable parameters (spine length, the largest acceptable angle for a new connection), largely reduces the manual input, and facilitates the cell quantification and diffusion simulations for the validation of biophysical modeling in MRI, such as the exchange effect in the brain gray matter [9-10].Acknowledgements
Research was supported by the Office of the Director of the NIH under award DP5 OD031854, and by the National Institute of Biomedical Imaging and Bioengineering (NIBIB) of the NIH under award U01 EB026996.References
1. Fieremans, E., & Lee, H. H. (2018). Physical and numerical phantoms for the validation of brain microstructural MRI: A cookbook. Neuroimage, 182, 39-61.
2. Shapson-Coe, A., Januszewski, M., Berger, D. R., Pope, A., Wu, Y., Blakely, T., ... & Lichtman, J. W. (2021). A connectomic study of a petascale fragment of human cerebral cortex. bioRxiv.
3. Tian, Q., Ngamsombat, C., Lee, H. H., Berger, D. R., Wu, Y., Fan, Q., ... & Huang, S. Y. (2020). Automated segmentation of human axon and myelin from electron microscopy data using deep learning for microstructural validation and simulation. In Proc. Int. Soc. Magn. Reson. Med. (p. 430).
4. Cuntz, H., Forstner, F., Borst, A., & Häusser, M. (2010). One rule to grow them all: a general theory of neuronal branching and its practical application. PLoS computational biology, 6(8), e1000877.
5. Li, J. R., Nguyen, V. D., Tran, T. N., Valdman, J., Trang, C. B., Van Nguyen, K., ... & Nguyen, T. M. P. (2019). SpinDoctor: a Matlab toolbox for diffusion MRI simulation. NeuroImage, 202, 116120.
6. Fang, C., Nguyen, V. D., Wassermann, D., & Li, J. R. (2020). Diffusion MRI simulation of realistic neurons with SpinDoctor and the Neuron Module. NeuroImage, 222, 117198.
7. Akram, M. A., Ljungquist, B., & Ascoli, G. A. (2021). Efficient metadata mining of web-accessible neural morphologies. Progress in Biophysics and Molecular Biology.
9. Palombo, M., Alexander, D. C., & Zhang, H. (2019). A generative model of realistic brain cells with application to numerical simulation of the diffusion-weighted MR signal. NeuroImage, 188, 391-402.
10. Jelescu, I. O., de Skowronski, A., Palombo, M., & Novikov, D. S. (2021). Neurite Exchange Imaging (NEXI): A minimal model of diffusion in gray matter with inter-compartment water exchange. arXiv preprint arXiv:2108.06121.9. Olesen, J. L., Østergaard, L., Shemesh, N., & Jespersen, S. N. (2021). Diffusion time dependence, power-law scaling, and exchange in gray matter. arXiv preprint arXiv:2108.09983.
Figures