4342
Self-supervised learning for multi-center MRI harmonization without traveling phantoms: application for cervical cancer classification
Xiao Chang1, Xin Cai1, Yibo Dan2, Yang Song2, Qing Lu3, Guang Yang2, and Shengdong Nie1
1the Institute of Medical Imaging Engineering, University of Shanghai for Science and Technology, Shanghai, China, 2the Shanghai Key Laboratory of Magnetic Resonance, Department of Physics, East China Normal University, Shanghai, China, 3the Department of Radiology, Renji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, China
1the Institute of Medical Imaging Engineering, University of Shanghai for Science and Technology, Shanghai, China, 2the Shanghai Key Laboratory of Magnetic Resonance, Department of Physics, East China Normal University, Shanghai, China, 3the Department of Radiology, Renji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, China
Synopsis
We proposed a self-supervised harmonization to achieve the generality and robustness of diagnostic models in multi-center MRI studies. By mapping the style of images from one center to another center, the harmonization without traveling phantoms was formalized as an unpaired image-to-image translation problem between two domains. The proposed method was demonstrated with pelvic MRI images from two different systems against two state-of-the-art deep-leaning (DL) based methods and one conventional method. The proposed method yields superior generality of diagnostic models by largely decreasing the difference in radiomics features and great image fidelity as quantified by mean structure similarity index measure (MSSIM).
Introduction
Multi-center MRI study becomes more and more important in AI-based medical imaging diagnosis. However, the contrast difference of multi-center images due to variations in scanners or scanning parameters has a negative effect on the generality of models trained on data from a few institutions. DL-based style transfer networks, such as U-Net[1], dualGAN[2] and cycleGAN [3, 4], have been widely used in Multi-center harmonization and achieved encouraging results. However, insufficient image fidelity when used without traveling phantoms remains a major impediment to the clinical applications of DL-based harmonization. In this study, we proposed a self-supervised style-transfer harmonization method to achieve reproducibility of multi-center studies [5] and achieved satisfying results on a two-center MRI dataset without traveling phantoms. Our method was a two-step transform consisting of a modified cycleGAN network for style transfer and a histogram matching module for structure fidelity.Methods
We retrospectively collected 116 pelvic MRI data scanned on two 3T MR systems from Renji Hospital. This retrospective study was approved by the institutional ethic committee and informed consent was waived. 75 cases were scanned on Philips Healthcare 3.0T Ingenia while 41 cases were scanned on GE Healthcare 3.0T Signa HDxt. We collected sagittal T2-weighted imaging (T2WI) images from picture archiving and communication systems (PACS). All regions of interest (ROI) of the cervical cancer were delineated manually with the ITK-SNAP software (http://www.itksnap.org) by a radiologist with 3 years’ experience in gynecological imaging. We resampled the images and ROIs of intra-slice resolutions below $$$0.65\times0.65mm^{2}$$$ or above $$$0.85\times0.85mm^{2}$$$ to $$$0.70\times0.70mm^{2}$$$. Then we rescaled the images to [0,1] followed by a dynamic normalization procedure. Finally, the images (sagittal slices) were cropped to $$$280\times280$$$. We used Philips’ images as the target of the style transfer. So, firstly, we used images from two systems to train a modified cycleGAN network (Figure 1) in an unpaired manner, to transfer Philips’ style to images from GE system. After images from GE system was transformed by the network, the generated image was paired with original GE image and input into a histogram matching module for strictly anatomical structure consistency. We compared the performance of our method with two state-of-the-art DL-based harmonization methods (dualGAN method [2], cycleGAN method [3]) and one traditional method (contrast limited adaptive histogram equalization, CLAHE [6]). We used Mann-Whitney U-test to find the radiomics features with a significant difference between two centers to prove the ability to remove inter-center difference. The mean structure similarity index measure (MSSIM [7]) and peak signal-to-noise ratio (PSNR) were used to quantify the quality of harmonized images. Downstream cervical cancer classification was used to demonstrate their contributions to the generality of diagnostic models. We used 75 Philips’ data (24 positive) as trainging cohort and 41 GE’s data (14 positive) as independent test cohort. And 5-fold cross validation was used on training cohort. The model information was listed in Table 1.Results
The Mann-Whitney U-test results were listed in Table 2. Through our method, the proportion of the features with a significant difference between images from two systems was reduced from 64/91 to 49/91 (p<0.05), indicating better harmonization among all competing methods (CLAHE: 54/91, dualGAN: 57/91, cycleGAN: 59/91). The numerical comparison (Table 3) of harmonized methods with PSNR and MSSIM between original MRI images and harmonized MRI images showed that the proposed method achieved the best performance on image fidelity and noise suppression. The Figure 2 was some image examples. We observed severe round artefacts (yellow arrows) using dualGAN and cycleGAN methods and hallucinated stripe-like artefacts in the background (green arrows) using CLAHE method, whereas improved image sharpness and structure fidelity (red insets) through the proposed algorithm. The clinical statistics of downstream classification were listed in Table 4. On the independent test cohort, our method achieved an AUC (0. 857) higher than those of CLAHE (0.770), dualGAN (0.828) and cycleGAN (0.812).Discussion and Conclusion
We proposed a self-supervised algorithm to transfer the style of images from one center to another center to achieve the generality and robustness of diagnostic models in multi-center MRI studies. By incorporating a histogram-matching module for image fidelity, the proposed algorithm performed data harmonization with minimal image structure loss. Compared previous studies [1, 8, 9] on image harmonization, the proposed method does not require travelling phantoms and makes fewer assumptions on the image characteristics. The major limitation of this work is that we only used a limited dataset of two systems in one institution to demonstrate our approach. In the future, more datasets involving more institutions should be used to further validate the proposed scheme. In conclusion, we proposed a self-supervised algorithm which used a cycleGAN-based style transfer network for style transfer and a histogram matching module for structure fidelity. The approach can be used to perform image harmonization without travelling phantom and without loss of image structures.Acknowledgements
No acknowledgement found.References
- Dewey, B.E., et al., DeepHarmony: A deep learning approach to contrast harmonization across scanner changes. Magn Reson Imaging, 2019. 64: p. 160-170.
- Zhong, J., et al., Inter-site harmonization based on dual generative adversarial networks for diffusion tensor imaging: application to neonatal white matter development. Biomed Eng Online, 2020. 19(1): p. 4.
- Modanwal, G., et al.
MRI image harmonization using cycle-consistent generative adversarial network.
in Medical Imaging 2020: Computer-Aided Diagnosis. 2020. International Society
for Optics and Photonics.
- Zhu, J.-Y., et al., Unpaired Image-to-Image Translation Using Cycle-Consistent Adversarial Networks. 2017 IEEE International Conference on Computer Vision (ICCV), 2017: p. 2242-2251.
- Ibrahim, A., et al., Radiomics for precision medicine: Current challenges, future prospects, and the proposal of a new framework. Methods, 2021. 188: p. 20-29.
- Singh, B.B. and S. Patel, Efficient medical image enhancement using CLAHE enhancement and wavelet fusion. International Journal of Computer Applications, 2017. 167(5): p. 0975-8887.
- Chow, L.S. and R. Paramesran, Review of medical image quality assessment. Biomedical signal processing and control, 2016. 27: p. 145-154.
- Koike, S., et al., Brain/MINDS beyond human brain MRI project: A protocol for multi-level harmonization across brain disorders throughout the lifespan. Neuroimage Clin, 2021: p. 102600.
- Kickingereder, P., et al., Radiomic subtyping improves disease stratification beyond key molecular, clinical, and standard imaging characteristics in patients with glioblastoma. Neuro-oncology, 2018. 20(6): p. 848-857.
Figures
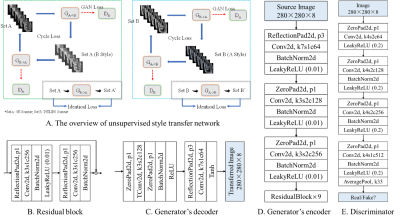
Figure 1 Our unsupervised
style transfer network (A) with both directions being trained simultaneously
(left: direction of A -> B (GE -> Philips), right: direction of B -> A
(Philips -> GE)) The generator is composed of three subnetworks: encoder (D)
for feature extraction, residual block (B) for stabilizing feature map, decoder
(C) for interpreting the transferred image from feature map. The GAN loss indicates adversarial loss. The cycle loss and identical
loss keeps image structures in an unpaired image-to-image translation.
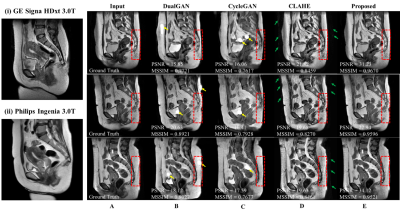
Figure 2 Harmonization results on 2D T2-weighted imaging (T2WI) sagittal slices. Example slices from two systems were shown in (i) and (ii). Column A showed a T2WI sagittal slice scanned on GE system. B, C, D, E showed harmonized images using dualGAN, cycleGAN, CLAHE and our proposed method, respectively. MSSIM and PSNR are annotated for each visualized slice.
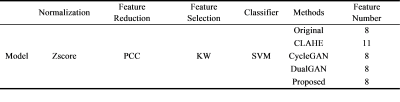
Table 1 The description of
the cervical cancer classification model.
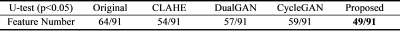
Table 2 The results of the
U-test: the proportion of features with a significant difference.

Table 3 Numerical
comparison (Mean±Std. Dev., 25%, 50% 75% and Max) of influence of different harmonization
algorithms to image structures, using PSNR and MSSIM (higher is better) between
original MRI images and harmonized MRI images.
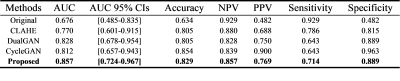
Table 4 The clinical
statistics of downstream classification on the independent test cohort comprised solely of
images from GE system. To demonstrate the effects of harmonization, the
classification model was trained using only images from Philips system.
DOI: https://doi.org/10.58530/2022/4342