4329
Automated IDH genotype prediction pipeline using multimodal domain adaptive segmentation (MDAS) model1Department of Electronic Science, Xiamen University, Xiamen, China, 2Department of Imaging, The First Affiliated Hospital of Fujian Medical University, Fuzhou, China, 3MSC Clinical & Technical Solutions, Philips Healthcare, ShenZhen, China
Synopsis
Mutation status of isocitrate dehydrogenase (IDH) in gliomas exhibits distinct prognosis. It poses challenges to jointly perform tumor segmentation and gene prediction directly using label-deprived multi-parametric MR images from clinics . We propose a novel multimodal domain adaptive segmentation (MDAS) framework, which derives unsupervised segmentation of tumor foci by learning data distribution between public dataset with labels and label-free targeted dataset. High-level features of radiomics and deep network are combined to manage IDH subtyping. Experiments demonstrate that our method adaptively aligns dataset from both domains with more tolerance toward distribution discrepancy during segmentation procedure and obtains competitive genotype prediction performance.
Introduction
Isocitrate dehydrogenase (IDH) mutant gliomas resembles wildtype in appearance, causing potential diagnostic trouble.1 Upon preoperative assessment of MR images, segmentation of tumor foci is an important step, which is almost supervised-learning oriented. On the other hand, most of IDH identification are based on radiomics or deep learning. Recent work extracted features by radiomics and support vector machine was used to identify gene mutations.2 Others employed hybrid deep learning model for tumor segmentation and IDH subtyping.3 These approaches again relied on manual labels. The unavailability of expert annotated labels prompt adoption of transfer learning to pretrain segmentation model using public data and then transfer it to target domain in absence of paired data.4 But such method demands complete consistent datasets, discrepancies induced by different imaging conditions would inevitably downgrade the performance. Domain adaptation such as CyCADA emphasizes consistency on semantic level to improve the outcome.6 Others claimed that image or feature alignment alone was not sufficient for cross-domain tasks, so unified network was proposed to coordinate the adaptive processes.7 Given specific clinical task, how to better retain pathologic information during unpaired transfer remains an open issue.Methods
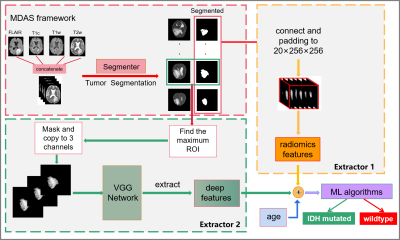
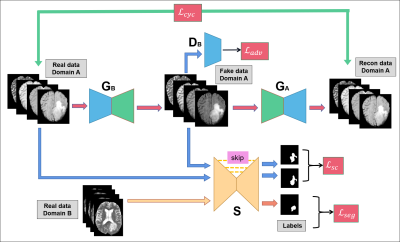
Our pipeline consists of two parts: unsupervised tumor segmentation via our proposed multimodal domain adaptive segmentation (MDAS) framework, extraction of radiomics and deep features to jointly predict IDH mutation status (Figure 1). The MDAS model (Figure 2) forms a cyclic structure with two generators and two discriminators. Both the input and output of the generator contain four channels, corresponding to four modalities of MR images being used. The network of discriminator has similar structure as CycleGAN,8 but with four input channels, which are compressed to one output patch to discriminate authenticity of the input. For segmentation module S, U-Net9 structure with residual connection is adopted. S is supervised trained by data with labels from Domain B, and it also predicts segmentation results of label-free data from Domain A post to domain transfer.Besides cycle consistency, we proposed segmentation consistency to additionally preserve semantic information of the lesion regions by imposing more constraints on generator. Errors induced by data distribution difference can be implicitly fed back to S to further enhance the segmentation performance. The objective function involving adversarial loss $$$\mathcal L_{adv}$$$, cycle consistency loss $$$\mathcal L_{cyc}$$$, segmentation loss $$$\mathcal L_{seg}$$$ and segmentation consistency loss $$$\mathcal L_{sc}$$$, is formulated as:
$$\mathcal L(G_A,G_B,D_A,D_B,S) =\mathcal L_{adv} (G_A,D_A)+\mathcal L_{adv} (G_B,D_B)+ λ\mathcal L_{cyc} (G_A,G_B)+L_{seg}(S)+γ\mathcal L_{sc} (G_A,S)(1)$$
For conciseness (see CycleGAN8 for loss with similar explanation), only $$$\mathcal L_{sc}$$$ is presented:
$$\mathcal L_{sc} (G_B,S) = \mathbb E_{x_A\sim p_d(x_A)} [\parallel S(G_B(x_A))-S(x_A)\parallel_1](2)$$
We use segmented results as ROIs when extracting radiomics features. The deep network feature extractor adopts VGG19 network10 as backbone with pre-trained ImageNet.11 We conduct embedded feature selection to obtain the most relevant features before training the random forest classifier.
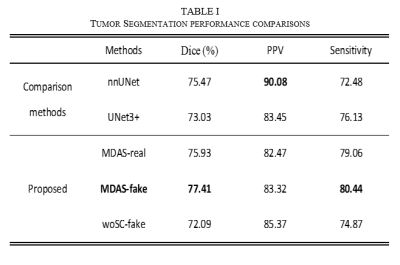
Grade II-IV gliomas of 110 cases (80 for training and 30 for test) with patient informed consent from our hospital were enrolled as data from target domain. BraTS2018 public dataset with annotated tumor labels were employed as data from source domain.12 Dice score was used to evaluate the segmentation outcome, positive predict value (PPV) and sensitivity were employed to respectively verify precision and recall rate of the model. We conducted comparative experiments with nnUNet13 and UNet3+14. The original data (MDAS-Real) and data after adaptive transfer (MDAS-fake) were also compared. Model without $$$\mathcal L_{sc}$$$ (MDAS-woSC) was compared as well. We also perform prediction with different features.
Results
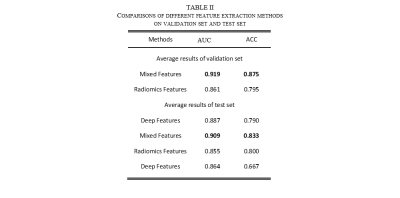
Figure 3 shows that MDAS obtains better segmented tumor foci. Table 1 shows our best model (MDAS-fake) outperforms nnUNet and UNet3+ in Dice by 1.9% and 4.4% respectively. The sensitivity reaches 80.44%, compared to 72.48% with nnUnet and 76.13% with UNet3+. Meanwhile, a decrease of PPV is observed. Compared to model using original data (MDAS-real) without going through multimodal adaptive transfer, the MDAS-fake post to transfer learning shows better performance. For experiment without adding $$$\mathcal L_{sc}$$$, Dice decreases by 5.32%. Table 2 depicts that mixed features from radiomics and deep network perform the best (AUC 0.909, ACC 0.833).Discussion
The main exploration goals of our work are, on one hand, to overcome problem of label scarcity; on the other hand, to combine advantages of deep learning with radiomics to extract more representative features. Domain adaptation transfer learning was used to train an optimized segmentation model, which automatically segment tumor foci for subsequent feature selection. And hybrid features of deep network and radiomics achieve better results than using either one alone. Besides, PPV and sensitivity have certain degree of mutual exclusion. In clinical practice, the cost of missed diagnosis is significantly higher than that of wrong diagnosis. Therefore, the sensitivity, which also increases fault tolerance of the model, might be given higher priority.Conclusion
We propose a multi-modal domain transfer pipeline for downstream task of subtyping IDH gene profiles. Our approach employs domain adaptation to narrow down distribution discrepancy across different datasets with multiple modalities, our label-free unsupervised learning largely eases the manual segmentation burden. The segmentation consistency loss better retains the lesion foci post to transfer. The adversarial property also makes our model readily extensive to scenarios where certain modality is missing or data suffering large modality differences.Acknowledgements
This work was supported in part by the National Natural Science Foundation of China under grant 82071913, 82071869, 82102021 and in part by the Joint Scientific Research Foundation of Fujian Provincial Education and Health Commission of China under grant 2019-WJ-10, and in part by Science and Technology Project of Fujian Province 2019Y0001.References
1. Smits M, Bent M. Imaging Correlates of Adult Glioma Genotypes. Radiology. 2017;284(2):316-331.
2. Peng H, Huo J, Li B, et al. Predicting Isocitrate Dehydrogenase (IDH) Mutation Status in Gliomas Using Multiparameter MRI Radiomics Features. J Magn Reson. 2020;53(5):1399-1407.
3. Chang K, Bai H, Zhou H, et al. Residual Convolutional Neural Network for the Determination of IDH Status in Low- and High-Grade Gliomas from MR Imaging. Clin Cancer Res. 2017;24:1073 - 1081.
4. Zhang Y, Miao S, Mansi T, et al. Task Driven Generative Modeling for Unsupervised Domain Adaptation: Application to X-ray Image Segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Granada. 2018;599-607.
5. Bousmalis K, Silberman N, Dohan D, et al. Unsupervised Pixel-Level Domain Adaptation with Generative Adversarial Networks. In: IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu. 2017;95-104.
6. Hoffman J, Tzeng E, Park T, et al. CyCADA: Cycle-Consistent Adversarial Domain Adaptation. In: International Conference on Machine Learning (ICML), Stockholm. 2018;1-15.
7. Chen C, Dou Q, Chen H, et al. Unsupervised Bidirectional Cross-Modality Adaptation via Deeply Synergistic Image and Feature Alignment for Medical Image Segmentation. IEEE Trans Med Imaging. 2020;39:2494-2505.
8. Zhu J-Y, Park T, Isola P, et al. Unpaired Image-to-Image Translation Using Cycle-Consistent Adversarial Networks. In: IEEE International Conference on Computer Vision (ICCV), Venice. 2017;2242-2251.
9. Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich. 2015;234-241.
10. Simonyan K, Zisserman A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In: International Conference on Learning Representations (ICLR), San Diego. 2015;1-14.
11. Deng J, Dong W, Socher R, et al. ImageNet: A large-scale hierarchical image database. In: IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR), Florida. 2009;248-255.
12. Menze B, Jakab A, Bauer S, et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans Med Imaging. 2015;34:1993-2024.
13. Isensee F, Kickingereder P, Wick W, et al. No New-Net. In: 4th International MICCAI Brainlesion Workshop, BrainLes 2018 Held in Conjunction with the Medical Image Computing for Computer Assisted Intervention Conference (MICCAI), Granada. 2018;234–244.
14. Huang H, Lin L, Tong R, et al. UNet 3+: A Full-Scale Connected UNet for Medical Image Segmentation. In: IEEE International Conference on Acoustics, Speech Signal Processing (ICASSP), Toronto. 2020;1055-1059.
Figures