4321
In vivo microstructural border delineation between areas of the human cerebral cortex using magnetic resonance fingerprinting (MRF) residuals1ARC training Centre for Innovation in Biomedical Imaging Technology, The University of Queensland, Brisbane, Australia, 2Centre for Advanced Imaging, The University of Queensland, Brisbane, Australia
Synopsis
We previously showed that magnetic resonance fingerprinting (MRF) residual signals can be used for in vivo voxel-wise parcellation of the human cerebral cortex, using supervised machine learning classification algorithms. However, previous work relied on brain atlases to provide probabilistic masks of cortical region to label samples to train a classification model. Here, we investigate the feasibility of developing automated atlas-free cortical border delineation in individuals. We demonstrate that 90% of the cortical border voxels identified by the proposed framework are co-localised with the borders between two cortical areas on the Juelich maximum probability map of cerebral cortex in six participants.
Introduction
In vivo accurate cortical parcellation of the human cerebral cortex is essential for understanding the association between structural and functional cortical variations,1 identifying the extent of the abnormal tissue accurately in neurosurgical applications2 and for examining microstructural changes due to progression of neurological disorders.3 The current resolution of in vivo MRI is several orders of magnitude larger than the scale of microstructural components in the brain tissue, requiring costly multi-contrast image acquisition and complex models for inferring microstructural features from in vivo MRI.4 Brain atlases are commonly used for identifying the extent of cortical regions on anatomical MRI of the individuals based on the average of histological analysis of postmortem tissue across a limited number of individuals.5, 6 Hence, only probabilistic maps are available which are not subject specific. We previously demonstrated that spatio-temporal MR fingerprinting7 residual signals8 contain rich descriptive area-specific representation for each voxel, leading to the development of a machine learning model for in vivo cortical parcellation.9 The study relied on a brain atlas to label the training samples.5 Here, we investigated the utility of area-specific information in MRF residuals to develop an automated atlas-free analysis pipeline for in vivo cortical border delineation in individuals.Methods
We acquired 1000 frames of 3D EPI-MRF scans from six healthy participants aged 31±4 years using a 7T MR scanner (Siemens Healthcare, Erlangen, Germany). MRF acquisition parameters were: flip angle= 10-77° (Figure 1a), TR= 41-99 ms (Figure 1b), TE= 12-48 ms (Figure 1b), Partial Fourier Phase= 6/8, voxel size= 1.4 × 1.4 × 1.4 mm3, and matrix size= 142 × 142 × 88. GRAPPA was used in phase and slice encoding directions. Chemical-shift-selective (CHESS) fat saturation was used to reduce EPI-specific artefacts.We generated an MRF dictionary using classical Bloch equation simulations: T1= 500-3500 ms (in steps of 10ms, 20ms, 30ms and 40ms for 500-1000ms, 1010-2010ms, 2010-3000ms, 3030-3500ms, respectively); T2*= 10-100 ms (in 2ms and 3ms increments for 10-60ms and 60-100ms, respectively); B1+= 0.4-1.2 in 0.05 increments). We then performed MRF dictionary matching and calculated MRF residual signal and the corresponding autocorrelation profile per voxel, as detailed previously.8
Our basis was that the differences between MRF residual signals would be smaller for voxels which are located in a single cortical region, compared to those across microstructurally different cortical borders. We used the maximum probability map of cortical areas extracted from the Juelich brain atlas5 to calculate the residual distance profiles along the cortical ribbon in each individual scan (see Figure 2).
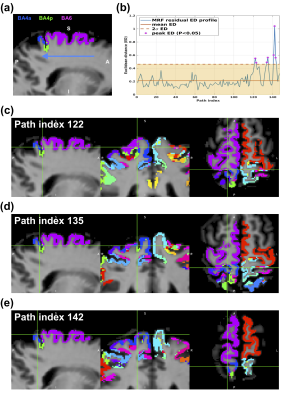
A total of 300 2D paths were extracted from sagittal, coronal and axial slices of 3D MRF images from six participants. The paths were used to measure the MRF residual differences between all adjacent voxels along the cortical ribbon and within the target areas. For each voxel along the path, we created a 3x3 2D kernel surrounding the voxel of interest, excluding the non-GM tissue. We then measured the average Euclidean distance (ED) between the MRF residual signal of the centre voxel and all other kernel voxels as we traversed each path. We then searched for the presence of peak distance values along each residual distance profile. The ED values outside two standard deviations from the mean distance values from each distance profile were considered as the peak distance values. The spatial location of the voxels corresponding to the peak ED values were examined to see if they were located within a 2-voxel radius from the border between any two cortical areas on the Juelich maximum probability map of the participant.
Results and Discussion
Figure 3a depicts an example path crossing the premotor area BA6 and primary motor areas BA4p and BA4a overlayed on an MRF scan from one participant, with residual distance profile for the path in Figure 3b. As an example, the spatial location of three of these peak voxels on the Juelich maximum probability map of the individual is illustrated in Figures 3c-e. We found that the first peak (at path indices 122-123 in Figure 3c) corresponds to the border between areas BA6 and BA4p on the Juelich maximum probability map. Additionally, the second (at path indices 134-15 in Figure 3d) and third (at path indices 141-13 in Figure 3e) peaks correspond to the border between BA4a and BA4p and between BA6 and BA4a, respectively. Across all participants, we observed that 90% of the border voxels detected were located at the 2-voxel radius of the border between two areas on the Juelich maximum probability map. The co-localisation of the peak ED values on the MRF residual distance profiles and the border voxels on the Juelich maximum probability map suggests the presence of microstructural information in the MRF residual signals, providing a tool for accurate in vivo voxel-wise cortical parcellation in individuals. This is in agreement with previously reported findings in an MRF residual-based cortical dissociation study.8Conclusion
Using MRF residual signals we demonstrated the feasibility of developing an automatic atlas-free border delineation between microstructurally distinct regions of the human cerebral cortex in vivo. This study sets the foundation for future work to develop robust unsupervised machine learning-based in vivo cortical parcellation in individuals.Acknowledgements
This research was conducted at the Australian Research Council Training Centre for Innovation in Biomedical Imaging Technology (IC170100035) and funded by the Australian Government. The authors also acknowledge the facilities and scientific and technical assistance of the National Imaging Facility, a National Collaborative Research Infrastructure Strategy (NCRIS) capacity, at the Centre for Advanced Imaging, The University of Queensland.
References
1. Wahl, M.; Li, Y. O.; Ng, J.; LaHue, S. C.; Cooper, S. R.; Sherr, E. H.; Mukherjee, P., Microstructural correlations of white matter tracts in the human brain. Neuroimage 2010, 51 (2), 531-541.
2. Awad, I. A.; Rosenfeld, J.; Ahl, J.; Hahn, J. F.; Luders, H., Intractable epilepsy and structural lesions of the brain: mapping, resection strategies, and seizure outcome. Epilepsia 1991, 32 (2), 179-86.
3. Rose, S. E.; Janke Phd, A. L.; Chalk, J. B., Gray and white matter changes in Alzheimer's disease: a diffusion tensor imaging study. Journal of Magnetic Resonance Imaging: An Official Journal of the International Society for Magnetic Resonance in Medicine 2008, 27 (1), 20-26.
4. Cercignani, M.; Bouyagoub, S., Brain microstructure by multi-modal MRI: Is the whole greater than the sum of its parts? Neuroimage 2018, 182, 117-127.
5. Amunts, K.; Mohlberg, H.; Bludau, S.; Zilles, K., Julich-Brain: A 3D probabilistic atlas of the human brain's cytoarchitecture. Science 2020, 369 (6506), 988-992.
6. Brodmann, K., Vergleichende Lokalisationslehre der Grosshirnrinde in ihren Prinzipien dargestellt auf Grund des Zellenbaues. Barth: 1909.
7. Ma, D.; Gulani, V.; Seiberlich, N.; Liu, K. C.; Sunshine, J. L.; Duerk, J. L.; Griswold, M. A., Magnetic resonance fingerprinting. Nature 2013, 495 (7440), 187-192.
8. Moinian, S.; Vegh, V.; O'Brien, K.; Reutens, D., Magnetic resonance fingerprinting residual signals can disassociate human grey matter regions. Brain Structure & Function 2021.
9. Moinian, S.; Vegh, V.; Reutens, D. In In vivo voxel-wise parcellation of the human cerebral cortex using 3D MR fingerprinting (MRF) and supervised machine learning classification, ISMRM, Virtual, Virtual, 2020.
Figures
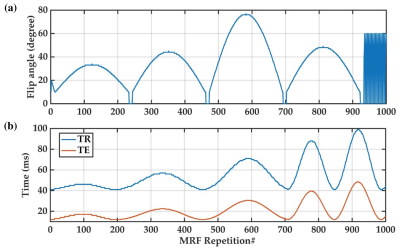
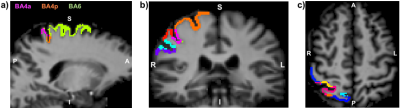
Figure 2 An example path along the cerebral cortex on a) sagittal, b) coronal and c) axial plane of the 3D MRF scans from one participant.