4311
Vascular Heterogeneity Model-based Deep Learning Reconstruction for High-Definition Dynamic Contrast Enhanced MRI1Department of Intelligent Precision Healthcare Convergence, Sungkyunkwan University, Suwon, Korea, Republic of, 2Department of Radiology, Seoul National University Hospital, Seoul, Korea, Republic of, 3Department of Biomedical Engineering, Sungkyunkwan University, Suwon, Korea, Republic of
Synopsis
To take characterization of various vascular contrast dynamics into account, in this work we propose a novel, vascular heterogeneity model based deep learning reconstruction from highly undersampled data for high-definition whole brain DCE MRI. To this end, we introduce a new, vascular contrast dynamics (VCD) weighted deep attention neural network (VACAN) consisting of: 1) a vascular adaptive attention 3D U-Net, 2) a multilayered non-negative matrix factorization (NMF) layer, and 3) a data consistency layer. Experimental studies are performed using highly undersampled patient data to validate the effectiveness of the proposed VACAN against conventional 3D U-Net.
Introduction
Dynamic contrast enhanced (DCE) MRI has been widely used in a clinical routine for non-invasive assessment of vascular structure and function1. In DCE MRI, vascular contrast dynamics is reflected in a time series of images, in which arteries and veins exhibit rapid, high uptake and washout of contrast agents (CA) while capillaries show relatively much slower uptake and washout behaviors. To take various contrast dynamics into account, in this work we propose a novel, vascular heterogeneity model based deep learning reconstruction from highly undersampled data for high-definition whole brain DCE MRI. To this end, we introduce a new, vascular contrast dynamics (VCD) weighted deep attention neural network (VACAN) consisting of: 1) a vascular adaptive attention 3D U-Net to accurately capture rapidly varying arterial signals while smoothing slowly varying microvascular signals, 2) a multilayered non-negative matrix factorization (NMF) layer to represent highly correlated microvascular signals in a low dimensional space for characterization of capillary tissues, and 3) a data consistency layer. Experimental studies are performed using highly undersampled patient data with brain tumors (R ~ 50) to validate the effectiveness of the proposed VACAN against conventional 3D U-Net.Materials and Methods
VACAN: Vascular Contrast Dynamics Weighted Deep Attention Neural NetworkTo accurately capture a rapidly changing portion of arterial contrast dynamics while tracing slowly varying capillary contrast dynamics with high fidelity, a mathematical model of the proposed method is constructed by:
$$L(W_{l},H_{l},X_{D}, P_{D})=\frac{1}{2}\mathrm{\left\| Y-F_{u}(P_{D}\odot X_{D}) \right\|}_{F}^{2}+\frac{\tau_{2}}{2}\sum_{l=1}^{N_{l}}\mathrm{\left\| M(X_{D}-W_{l}H_{l})\right\|}_{F}^{2}+\frac{\lambda_{2}}{2}\mathrm{\left\| X_{D}-\textbf{DNN}(MX_{D})\right\|}_{F}^{2} \quad s.t. W_{l}\geqslant 0,H_{l}\ge 0$$
Where $$$\textbf{DNN}(·)$$$ is the VCD-weighted Attention 3D U-Net for vascular adaptive processing; $$$\sum_{l=1}^{N_{l}}\mathrm{\left\| M(X_{D}-W_{l}H_{l})\right\|}_{F}^{2}$$$ is a spatially weighted multilayered NMF; $$$\mathrm{\left\| Y-F_{u}(P_{D}\odot X_{D}) \right\|}_{F}^{2}$$$ is the data consistency.
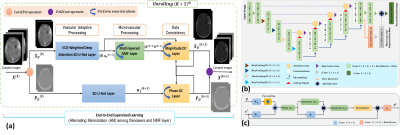
The objective function $$$L$$$ is minimized using an alternating minimization method using the vascular contrast dynamics weighted deep attention neural network. Fig. 1 shows the architecture and key components of Vascular Contrast Dynamics Weighted Deep Attention Neural Network - VACAN: (a)VACAN uses complex-valued undersampled images estimated by a sensitivity encoding operator as inputs. Then, the inputs alternate among vascular adaptive, microvascular processing, and data consistency. (b)The multi-task VCD-weighted Attention 3D U-Net. (c)A proposed Spatio-temporal weighted attention mechanism.
Data acquisition and preprocessing
We acquired a time series of 3D whole-brain DCE data in 13 patients with brain tumors using our customized 3D GRE pulse sequence2 on a 3T whole-body MR scanner (Skyra, Siemens Healthineers, Erlangen, Germany). Each set of 3D data in a single time frame was vastly undersampled on the Cartesian grid (ky-kz) in a pseudo-golden-angle radial scheme3 roughly with a reduction factor (R) of 50. Imaging parameters were as follows: TR/TR = 3.18/1.1 ms, flip angle= 15°, matrix size = 192 × 192 × 144, readout bandwidth = 650 Hz/Px, number of time frames = 170, spatial resolution = 1.0mm3, temporal resolution = 1.5 sec, and imaging time = 4.5 min. Additionally, variable flip angle imaging4 was performed prior to DCE MRI using 2°, 8°, and 15° to estimate a reference T10 map for quantitative analysis. All experiments were performed under the approval of the Institutional Review Board (IRB no.: 1806-137-954) and written informed consents were obtained from all patients prior to imaging.
Training, Validation, Testing
We divided noise-augmented data into training, validation, and test datasets. The training dataset contains 137 slices from 9 patients; the validation dataset contains 3 slices from 1 patient; test dataset contains 97 slices from 3 patients. Intermediate to high frequency signals in 3 neighboring time frames were shared, yielding R~21 for image reconstruction. Coil sensitivity maps were calculated using the ESPIRIT5 with full-sampled data in the central k-space. Images were combined in the coil direction using the sensitivity encoding operator. VACAN was trained by an Adam optimizer with an epoch ~ 300. An unrolling step was set to 2. The trainable parameters of regularization $$${\lambda_P,\lambda_2,\tau_2}$$$ were initialized to $$${{3e}^{-2},{3e}^{-2},{1e}^{-2}}$$$. The proposed VACAN was implemented using Tensorflow 1.15.0 installed on Ubuntu 18.04 LTS.
Results
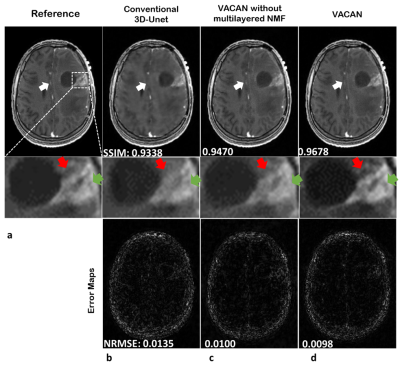
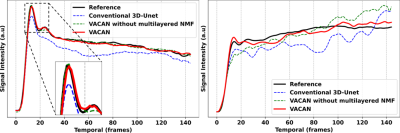
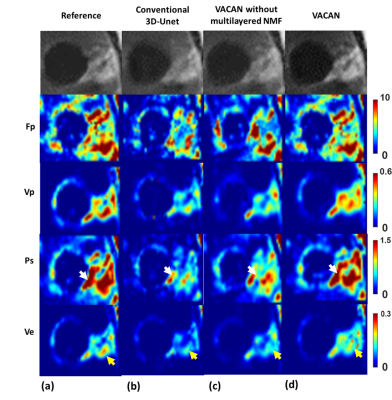
Fig. 2 compares DCE images reconstructed using the reference (a), conventional 3D U-Net (b), the VACAN without the multilayered NMF (c), and the VACAN (d). Compared with the reference, conventional 3D U-Net exhibits severe blurring and contrast loss (white arrow) while the VACAN with/without the multilayered NMF restores image sharpness and heterogeneous tumor structures. Note that adding the multilayer NMF to the VACAN enhances delineation of tumor heterogeneities (green and red arrows). Fig. 3 shows the corresponding temporal signal evolutions reconstructed using the above three methods. The VACAN with/without the multilayered NMF delineates arterial contrast dynamics accurately (Fig. 3a). Additionally, the multilayered NMF inside the proposed VACAN enhances fidelity of capillary contrast dynamics to that of the reference (Fig. 3b). Fig. 4 compares tracer-kinetic (TK) parameter maps (4-parameter 2CX model; $$$F_{p}$$$: plasma flow, $$$v_{p}$$$: plasma volume fraction, $$$PS$$$: permeability-surface area product, $$$v_{e}$$$: interstitial volume fraction). The proposed VACAN yields TK maps very close to the reference, outperforming the other methods including conventional 3D U-Net.Discussion and Conclusion
In conclusion, we successfully demonstrated the feasibility of the proposed VACAN based deep learning reconstruction from highly undersampled data (R~ 50) to achieve high-definition whole brain DCE MRI. The proposed VACAN outperforms competing methods regarding evaluation metrics (SSIM, NRMSE) and quantitative map accuracies. Additionally, whole brain data was reconstructed roughly in 3 minutes.Acknowledgements
This work is supported by NRF-2018M3C7A, KMDF-202011B35, and KMDF-202011C20References
1. S. Park, E. Y. Kim, C.-H. Sohn, and J. Park, “Dynamic contrast-enhanced mr angiography exploiting subspace projection for robust angiogram separation,” IEEE Transactions on Medical Imaging, 2016.
2. Joon Sik Park, Seung-Hong Choi, Chul-Ho Sohn, and Jaeseok Park, “Joint Reconstruction of Vascular Structure and Function Maps in Dynamic Contrast Enhanced MRI Using Vascular Heterogeneity Priors,” IEEE Transactions on Medical Imaging, 2021.
3. Li Feng,Robert Grimm,Kai Tobias Block,Hersh Chandarana,Sungheon Kim,Jian Xu,Leon Axel,Daniel K. Sodickson,Ricardo Otazo, “Golden-angle radial sparse parallel MRI: Combination of compressed sensing, parallel imaging, and golden-angle radial sampling for fast and flexible dynamic volumetric MRI,” Magnetic Resonance in Medicine, 2013.
4. S. C. Deoni, T. M. Peters, and B. K. Rutt, “High resolution T1 and T2 mapping of the brain in a clinically acceptable time with DESPOT1 and DESPOT2,” Magnetic Resonance in Medicine, 2005.
5. Martin Uecker,Peng Lai,Mark J. Murphy,Patrick Virtue,Michael Elad,John M. Pauly,Shreyas S. Vasanawala,Michael Lustig, “ESPIRiT—an eigenvalue approach to autocalibrating parallel MRI: Where SENSE meets GRAPPA,” Magnetic Resonance in Medicine, 2013.
Figures