4309
Susceptibility Weighted MRI for Predicting Critical Developmental Regulatory S100 Proteins in Meningiomas at 3T1Institute of Biomedical Engineering, Bogazici University, Istanbul, Turkey, 2Department of Radiology, Basaksehir Cam and Sakura City Hospital, Istanbul, Turkey, 3Electric and Electronic Engineering Department, Bogazici University, Istanbul, Turkey, 4Department of Medical Pathology, Acibadem Mehmet Ali Aydinlar University, Istanbul, Turkey, 5Center for Neuroradiological Applications and Reseach, Acibadem Mehmet Ali Aydinlar University, Istanbul, Turkey, 6Health Institutes of Turkey, Istanbul, Turkey, 7Department of Medical Engineering, Acibadem Mehmet Ali Aydinlar University, Istanbul, Turkey, 8Department of Neurosurgery, Acibadem Mehmet Ali Aydinlar University, Istanbul, Turkey, 9Department of Radiology, Acibadem Mehmet Ali Aydinlar University, Istanbul, Turkey
Synopsis
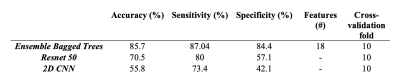
Meningiomas are the most common primary intracranial tumors in adults. S100 protein expression (S100+) in meningiomas is a marker of neural crest origin. Eighty-four patients with preoperative MRI were included in this IRB approved study. The whole tumor volumes were segmented from FLAIR, followed by co-registration onto SWI. Supervised machine and deep learning methods were employed to categorize meningiomas into S100+ and S100- groups based on SWI histogram values. Ensemble bagged trees resulted in an accuracy of 85.7% (sensitivity=87.0 % and specificity=84.4 %), while a Resnet 50 architecture had 70.5% accuracy (sensitivity=80%, specificity=57.1%) for predicting S100 protein expression in meningiomas.
Introduction
Meningiomas are the most frequent primary intracranial tumors in adults that typically emerges at the arachnoid or dural border cells [1, 2]. Low-grade meningiomas (World Health Organization (WHO) grade 1), which is the most common type, can be followed for a long period of time [3]; whereas, the more aggressive types (WHO grades 2 and 3) should be treated with early and complete resection due to their higher recurrence rate and invasive behavior [4]. Aggression of meningiomas varies according to demographic, anatomical, histopathological and clinical variables, which is thought to be due to differences in the expression of developmental regulatory proteins [5-7]. The S100 protein, a critical growth regulator, is used to identify tumors originating from the neural crest and is primarily expressed in the early stages of differentiation [8]. Even though, magnetic resonance imaging (MRI) and molecular markers are routinely utilized to make a preoperative diagnosis, surgical excision of the tumor is required for histological confirmation. Hence, preoperative evaluation of S100 positivity of meningioma might help with treatment decision-making. Susceptibility weighted MRI (SWI) facilitates understanding of tumor aggressiveness in meningiomas by providing information about tumor vasculature, intratumoral calcification, and microhemorrhage. This study aims to evaluate SWI findings in predicting S100 protein expression in meningiomas using machine learning and deep learning models.Methods
Eighty-four patients with meningioma (32M/52F, mean age= 49.7±13.4 years), who underwent a preoperative MRI, were retrospectively included in this IRB approved study. All patients provided written informed consent. Tumor samples underwent histopathological examination and immunohistochemical analysis using anti S-100 antibody to obtain S100 protein expression. MRI protocol consisted of FLAIR (TR/TE=8320/92ms, TI =2000ms, field of view (FOV) =220mm, slice thickness=3mm) and SWI (TR/TE=28/20ms, FOV=220mm, slice thickness=1.6mm) on a 3T clinical MR scanner (Siemens Healthcare, Erlangen, Germany). The MR system automatically generated magnitude and phase images and final SWI. The whole tumor volumes were segmented on FLAIR images using 3D Slicer version 4.8.1 (http://slicer.org/). The FLAIR images of each subject were registered to the high resolution SWI using FMRIB Software Library (FSL; http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/). Afterwards, the obtained transformation matrix was applied to register the segmentation masks onto the SWI for each subject. First-order textural features of SWI within FLAIR hyperintensity were extracted, and min-max normalization was applied in MATLAB (MathWorks Inc., Natick, MA, USA). A Mann-Whitney U test was employed to assess the SWI feature differences between S100 negative (S100-) and positive (S100+) groups. Histopathologic subtypes were compared among the S100+ and S100- groups using a Fischer’s exact test. Machine learning classification was conducted using morphologic and SWI features of tumors, and clinical information of patients including age, sex, growth pattern, localization, peritumoral edema, sinus invasion, hyperostosis, bone destruction and intratumoral calcification in MATLAB Statistics and Machine Learning Toolbox R2019b. Naive random over-sampling method was used for overcoming the class imbalance problem (9). Finally, convolution neural networks (CNN) and transfer learning approaches along with data augmentation were employed to categorize SWI of meningiomas into S100+ and S100- groups.Results
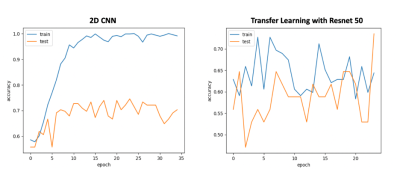
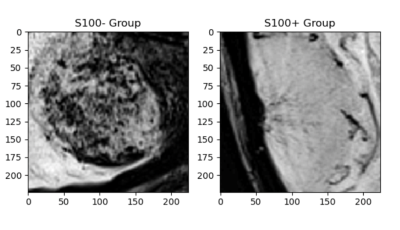
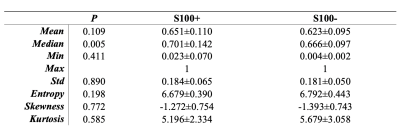
Twenty-eight meningiomas were S100+ and 56 patients were S100-. Fibrous/transitional meningioma subtype distribution was significantly higher in S100+ group than S100- group (P<0.001). The median SWI values within the volume of interest were significantly higher in S100+ group (normalized pixel value=0.701±0.142) than the S100- group (normalized pixel value=0.666±0.097) (P =0.005); whereas entropy, skewness and kurtosis values were similar between these two groups (Table 1). Ensemble bagged trees had the highest accuracy of 85.7% (sensitivity=87.0 % and specificity=84.4 %) to categorize S100 molecular subsets based on SWI, clinical and morphological features (Table 2). The prediction performance of 2D CNN and transfer learning (Resnet 50) methods were lower than that of the classical machine learning methods (Figures 1 and 2). While the transfer learning approach (Resnet 50) had a higher accuracy of 70.5% (sensitivity=80%, specificity=57.1%); 2D CNN resulted in an accuracy of only 55.8% (sensitivity=73.4%, specificity=42.1%) for predicting S100 molecular subsets in this small patient cohort (Table 2).Discussion
Our study demonstrated that SWI might be helpful for the prediction S100 protein positivity in meningiomas, using machine learning methods. Immunohistochemical analysis of S100 protein expression is routinely used for pathological differential diagnosis. S100 protein positivity is a marker for neural crest derived meningiomas, since S100 protein is related to early state of differentiation in neural crest cell (8, 10, 11). Meningiomas with S100 protein expression have a tendency for a lower WHO grade, which is in line with the findings of higher SWI values and more fibrous/transitional histopathological subtype in S100+ group than S100- group (12-14). Moreover, lower SWI values in S100- group might be related to tumor vascularity and the tumor grade.Conclusion
Machine learning algorithms applied on SWI might help with revealing underlying developmental regulator protein expression in meningiomas, which might offer potential guidance in treatment decision-making. Future studies will validate machine learning results in a larger patient cohort.Acknowledgements
This study was supported by the Scientific and Technological Research Council of Turkey (TUBITAK) grant 119S520.References
1. Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathologica. 2016;131(6):803-20.
2. Yamashima T. Human Meninges: Anatomy and Its Role in Meningioma Pathogenesis. In: Lee JH, editor. Meningiomas. London: Springer London; 2009. p. 15-24.
3. Rogers L, Barani I, Chamberlain M, Kaley TJ, McDermott M, Raizer J, et al. Meningiomas: knowledge base, treatment outcomes, and uncertainties. A RANO review. J Neurosurg. 2015;122(1):4-23.
4. Black PM, Villavicencio AT, Rhouddou C, Loeffler JS. Aggressive surgery and focal radiation in the management of meningiomas of the skull base: preservation of function with maintenance of local control. Acta Neurochir (Wien). 2001;143(6):555-62.
5. Clark VE, Harmancı AS, Bai H, Youngblood MW, Lee TI, Baranoski JF, et al. Recurrent somatic mutations in POLR2A define a distinct subset of meningiomas. Nature Genetics. 2016;48(10):1253-9.
6. Sahm F, Schrimpf D, Stichel D, Jones DTW, Hielscher T, Schefzyk S, et al. DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. The Lancet Oncology. 2017;18(5):682-94.
7. Harmancı AS, Youngblood MW, Clark VE, Coşkun S, Henegariu O, Duran D, et al. Integrated genomic analyses of de novo pathways underlying atypical meningiomas. Nature Communications. 2017;8(1):14433.
8. Donato R, Cannon BR, Sorci G, Riuzzi F, Hsu K, Weber DJ, et al. Functions of S100 proteins. Curr Mol Med. 2013;13(1):24-57.
9. Menardi G, Torelli N. Training and assessing classification rules with imbalanced data. Data Mining and Knowledge Discovery. 2014;28(1):92-122.
10. Ng J, Celebre A, Munoz DG, Keith JL, Karamchandani JR. Sox10 is superior to S100 in the diagnosis of meningioma. Appl Immunohistochem Mol Morphol. 2015;23(3):215-9.
11. Ulgen E, Bektasoglu PK, Sav MA, Can O, Danyeli AE, Hizal DB, et al. Meningiomas Display a Specific Immunoexpression Pattern in a Rostrocaudal Gradient: An Analysis of 366 Patients. World Neurosurg. 2019;123:e520-e35.
12. Al-Rashed M, Foshay K, Abedalthagafi M. Recent Advances in Meningioma Immunogenetics. Front Oncol. 2019;9:1472.
13. Nassiri F, Liu J, Patil V, Mamatjan Y, Wang JZ, Hugh-White R, et al. A clinically applicable integrative molecular classification of meningiomas. Nature. 2021;597(7874):119-25.
14. Hancq S, Salmon I, Brotchi J, Gabius HJ, Heizmann CW, Kiss R, et al. Detection of S100B, S100A6 and galectin-3 ligands in meningiomas as markers of aggressiveness. Int J Oncol. 2004;25(5):1233-40.
Figures