4302
XCloud-pFISTA: A Medical Intelligence Cloud for Accelerated MRI Reconstruction1Biomedical Intelligent Cloud R&D Center, Department of Electronic Science, National Institute for Data Science in Health and Medicine, Xiamen University, Xiamen, China, 2School of Computer and Information Engineering, Xiamen University of Technology, Xiamen, China, 3Department of Instrumental and Electrical Engineering, Xiamen University, Xiamen, China, 4Biomedical Intelligent Cloud R&D Center, School of Electronic Science and Engineering, China Mobile Group, Xiamen, China
Synopsis
Machine learning and artificial intelligence have shown remarkable performance in accelerated magnetic resonance imaging (MRI). Cloud computing technologies have great advantages in building an easily accessible platform to deploy advanced algorithms. In this work, we develop a high-performance medical intelligence cloud computing platform (XCloud-pFISTA) to reconstruct MRI images from undersampled k-space data. Two state-of-the-art approaches of the Projected Fast Iterative Soft-Thresholding Algorithm (pFISTA) family have been successfully implemented on the cloud. This work can be considered as a good example of cloud-based medical image reconstruction and may benefit the future development of integrated reconstruction and online diagnosis system.
Purpose
Magnetic resonance imaging (MRI) is a significant clinical diagnosis tool but suffers from relatively long data acquisition time. To achieve fast imaging, sparse sampling and parallel imaging are hot topics for decades1-7. Optimization methods and deep learning have raised great concerns for accelerating imaging speed and improving image quality. Although both of them have excellent performance, the environment configuration is complex and cumbersome. To the best of our knowledge, there are few integrated and easy-to-use platforms for MRI researchers to easily and effectively do the medical image processing and analysis. Cloud computing is a data-centered distributed computing mode8, 9. It is accessible, high-integrated, safe and has virtualized hardware. Compared with traditional computing mode, it has superior technologies in data storage and computing power. In this work, we developed XCloud-pFISTA, an easy-to-use, high-performance, and open access medical intelligent cloud for accelerated MRI. So far, two representative MRI reconstruction approaches (optimization method pFISTA10 and deep learning method pFISTA-Net11) have been implemented on the cloud. Users can enjoy these advanced approaches to conveniently and efficiently recover high-quality images from partial sampled k-space.Method
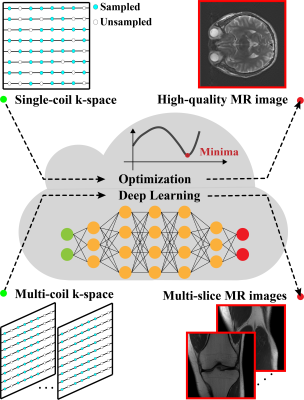
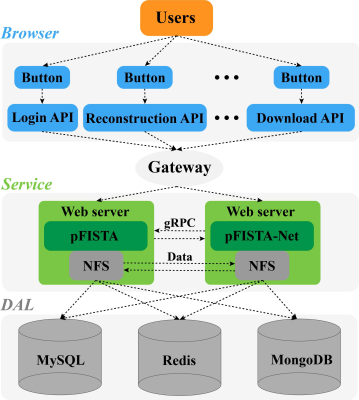
MRI reconstruction approaches: pFISTA is an iterative approach for single-coil MRI reconstruction, which employs sparsity of images under the redundant representation of tight frame10. Compared with other state-of-the-art approaches, pFISTA has the advantages of faster convergence speed, insensitivity to step size, and only needs to set one parameter. pFISTA-Net11 is an unrolled deep neural network for parallel MRI reconstruction. In pFISTA-Net, manual-craft sparsity transform is replaced by learnable convolutional filters, making it more robust for reconstructing images in different scenarios. The residual structure is employed to improve the learning capabilities of the network. As shown in Figure 1, XCloud-pFISTA that integrates above-mentioned two approaches will make them easier for using by MRI researchers, and perform further medical image analysis.System architecture: Cloud computing is a kind of distributed computing, which is accessible, high-integrated, safe, and has virtualized hardware12,13. Figure 2 shows the entire system architecture of XCloud-pFISTA. It adopts the browser/service (B/S) architecture, mainly consists of three parts: browser, service, and data access layer (DAL). Browser layer including some visual api buttons. Service layer including nginx transport, distributed web servers and use Google remote procedure call (gRPC) to communicate with python algorithms, Network file system (NFS) to ensure data consistency. Data access layer using redis, mysql or mangoDB to storage platform data.
Results
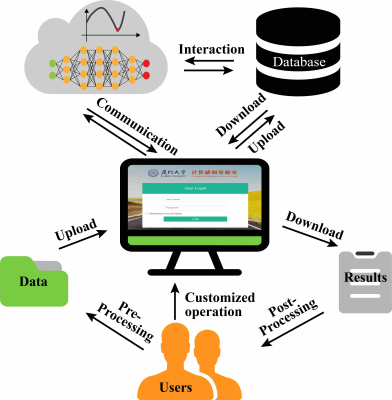
XCloud-pFISTA workflow: Up to now, XCloud-pFISTA supports single-coil and multi-coil MRI reconstruction, and is open access at http://36.134.50.123:2345 (Test account: ISMRM, Password: ISMRM2021). Figure 3 shows the whole workflow of the usage of XCloud-pFISTA.Reconstruction results and comparison: we compare the cloud and local results. Both of pFISTA and pFISTA-Net are written in Python. The parameters used in pFISTA10 and pFISTA-Net11 are all consistent with original papers. Here, Local reconstruction is conducted on a Windows PC with 8 cores CPU, 8 GB RAM, and one NVIDIA GTX 850M. For reconstruction on cloud, we use the China Mobile e-cloud host with 16 cores CPU, 128 GB RAM, and two NVIDIA T4 GPUs. For quantitative comparison, we adopt the relative l2 norm error (RLNE) defined as
\begin{equation} \mathbf{RLNE=} \frac{\left \|\mathbf{x}-\mathbf{\hat{x}}\right \| _{2} }{\left \|\mathbf{x}\right \|_{2} } \tag{1} \end{equation}
where $$$\mathbf{x}$$$ is the ground truth and $$$\mathbf{\hat{x} }$$$ is the reconstructed imaged.
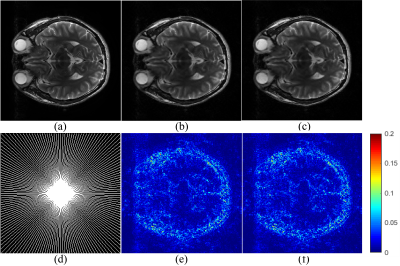
For single-coil data, pFISTA is used to reconstruct the T2-weighted brain dataset, which is acquired from a healthy volunteer at a 3T Siemens Trio Tim MRI scanner with 32-coils using a T2-weighted turbo spin echo sequence (TR/TE = 6100/99 ms, FOV = 220×220 mm2, slice thickness = 3mm). Results of pFISTA on local and cloud are shown in Figure 4. For multi-coil data, pFISTA-Net is used to reconstruct the knee dataset from a public knee dataset14: coronal proton-density (15 knee coils, TR/TE = 2750/27 ms, FOV = 320×320 mm2, In-plane resolution = 0.49×0.44 mm2). Results of pFISTA-Net on local and cloud are shown in Figure 5. Figure 4 and Figure 5 show that XCloud-pFISTA obtains the same high-quality results as on local, while the reconstruction time is reduced remarkably. Thus, the results suggest that, XCloud-pFISTA is a reliable and high-performance cloud computing platform for accelerated MRI.
Conclusion
In summary, we developed XCloud-pFISTA, a reliable, high-performance, and open access medical intelligence cloud for accelerated MRI. Two state-of-the-art approaches, pFISTA and pFISTA-Net, have been implemented on cloud for single-coil and multi-coil image reconstruction. We are constantly improving the XCloud-pFISTA and hope to provide an advanced cloud computing platform, which integrates numerous state-of-the-art approaches for MRI processing and analysis. Furthermore, we also plan to integrate artificial intelligence diagnostics on XCloud-pFISTA in the future.Acknowledgements
This work was supported in part by National Natural Science Foundation of China (62122064, 61971361, 61871341, 61811530021, and U1632274), National Key R&D Program of China (2017YFC0108703), Xiamen University Nanqiang Outstanding Talents Program, China Mobile Communications Group Fujian Co,. Ltd. Xiamen Branch Project (XDHT2021004C), Natural Science Foundation of Fujian Province of China under Grant 2021J011184.
The correspondence should be sent to Prof. Xiaobo Qu (Email: quxiaobo@xmu.edu.cn)
References
[1] M. Lustig, D. Donoho, and J. M. Pauly, “Sparse MRI: The application of compressed sensing for rapid MR imaging,” Magn. Reson. Med., vol. 58, no. 6, pp. 1182-1195, 2007.
[2] K. P. Pruessmann, M. Weiger, M. B. Scheidegger, and P. Boesiger, “SENSE: Sensitivity encoding for fast MRI,” Magn. Reson. Med., vol. 42, no. 5, pp. 952-962, 1999.
[3] M. Uecker, P. Lai, M. J. Murphy, P. Virtue, M. Elad, J. M. Pauly, S. S. Vasanawala, and M. Lustig, “ESPIRiT—an eigenvalue approach to autocalibrating parallel MRI: Where SENSE meets GRAPPA,” Magn. Reson. Med., vol. 71, no. 3, pp. 990-1001, 2014.
[4] M. A. Griswold, P. M. Jakob, R. M. Heidemann, M. Nittka, V. Jellus, J. Wang, B. Kiefer, and A. Haase, “Generalized autocalibrating partially parallel acquisitions (GRAPPA),” Magn. Reson. Med., vol. 47, no. 6, pp. 1202-1210, 2002.
[5] Z. Liang, “Spatiotemporal imaging with partially separable functions,” IEEE International Symposium on Biomedical Imaging (ISBI), pp. 988-991, 2007.
[6] Z. Zhan, J. Cai, D. Guo, Y. Liu, Z. Chen, and X. Qu, “Fast multiclass dictionaries learning with geometrical directions in MRI reconstruction,” IEEE Trans. Biomed. Eng., vol. 63, no. 9, pp. 1850-1861, 2016.
[7] X. Qu, Y. Hou, F. Lam, D. Guo, J. Zhong, and Z. Chen, “Magnetic resonance image reconstruction from undersampled measurements using a patch-based nonlocal operator,” Med. Image Anal., vol. 18, no. 6, pp. 843-856, 2014.
[8] M. Armbrust, A. Fox, R. Griffith, A. D. Joseph, R. Katz, A. Konwinski, G. Lee, D. Patterson, A. Rabkin, and I. Stoica, “A view of cloud computing,” Commun. ACM, vol. 53, no. 4, pp. 50-58, 2010.
[9] T. Dillon, C. Wu, and E. Chang, “Cloud computing: Issues and challenges.” IEEE International Conference on Advanced Information Networking and Applications (AINA), pp. 27-33, 2010.
[10] Y. Liu, Z. Zhan, J. Cai, D. Guo, Z. Chen, and X. Qu, “Projected iterative soft-thresholding algorithm for tight frames in compressed sensing magnetic resonance imaging,” IEEE Trans. Med. Imaging, vol. 35, no. 9, pp. 2130-2140, 2016.
[11] T. Lu, X. Zhang, Y. Huang, D. Guo, F. Huang, Q. Xu, Y. Hu, L. Ou-Yang, J. Lin, Z. Yan, and X. Qu, “pFISTA-SENSE-ResNet for parallel MRI reconstruction,” J. Magn. Reson., vol. 318, pp. 106790, 2020.
[12] F. Zhao, C. Li, and C. F. Liu, “A cloud computing security solution based on fully homomorphic encryption,” International Conference on Advanced Communication Technology (ICACT), pp. 485-488, 2014.
[13] Y. Jadeja, and K. Modi, “Cloud computing-concepts, architecture and challenges,” International Conference on Computing, Electronics and Electrical Technologies (ICCEET), pp. 877-880, 2012.
[14] K. Hammernik, T. Klatzer, E. Kobler, M. P. Recht, D. K. Sodickson, T. Pock, and F. Knoll, “Learning a variational network for reconstruction of accelerated MRI data,” Magn. Reson. Med., vol. 79, no. 6, pp. 3055-3071, 2018.
Figures