4208
Deep Learning Method Based Automatic Subtraction of Liver DCE-MR Images1The department of medical imaging, Sun Yat-sen University,Cancer Center, Guangzhou, China, 2States key laboratory in South China, Guangzhou, China, 3Shukun (Beijing) Technology Co., Ltd, Beijing, China, 4Center for Biomedical Imaging Research, Department of Biomedical Engineering,School of Medicine, Tsinghua University, Beijing, China
Synopsis
Subtraction images are an important part of routine multi-phase contrast-enhanced MRI for characterizing enhancement of lesions which are intrinsically hyperintense on T1-weighted imaging. Successful subtraction MRI is dependent on precise 3D co-registration of the pre- and contrast-enhanced source data. However, there still lack a robust, convenient, time efficient and labor free method for automatically image subtraction. This study developed a deep learning based nonrigid registration algorithm, measure the improvement in displacement after registration using anatomic landmarks and automatically generate the subtraction liver images.
Introduction
Subtraction images are an important part of routine multi-phase contrast-enhanced MRI for characterizing enhancement of lesions which are intrinsically hyperintense on T1-weighted imaging [1]. One crucial role for subtraction imaging is the detection of enhancement in the diagnosis and treatment response evaluation of focal hepatic lesions, such as HCCs [2].Successful subtraction MRI is dependent on precise 3D image co-registration of the pre- and contrast-enhanced source data. Misregistration as a result of patient motion artifact (e.g., breathing, heartbeat, and bowel peristalsis) can lead to suboptimal subtraction or even failure of the subtraction process.Various image registration techniques have been described for contrast-enhanced liver MRI, and convolutional neural networks (CNN) have been used in deformable registration tasks [3,4]. The purpose of this study was to develop a deep learning based nonrigid registration algorithm, measure the improvement in displacement after registration using anatomic landmarks and automatically generate the subtraction liver images.Methods
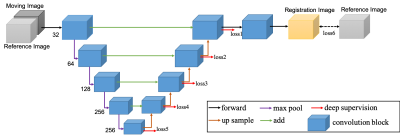
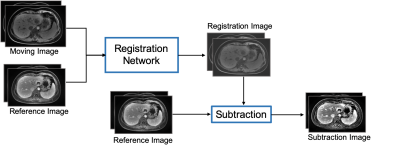
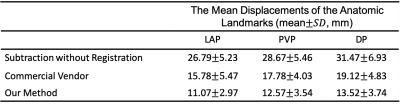
A retrospective study included multiphase DCE MR images from 124 patients were included to build and validate the model. An external test cohort including images from 96 patients was included to further verify the generalization capability of the proposed model.The pre-enhanced images (moving images) are affine registered to contrast-enhanced images (reference images, e.g., arterial, portal venous, and delayed) using a deeply supervised 3D U-net liver registration algorithm (Figure 1). The deeply supervised 3D U-net architecture is a neural network consisted of the encoder and decoder part [5]. In our network, all convolutional layers use kernel size of 3×3×3 and stride of 2, except the last one uses 1×1×1 kernel size and strides of 1. In the convolutional blocks, Batch normalization (BN) and Rectified linear unit (ReLU) are adopted to speed up the training and to enhance the gradient back propagation. After image registration, registered pre-enhanced images were subtracted from contrast-enhanced images to create the subtraction images, as shown in Figure 2.Ten anatomic landmarks, as shown in Table 1, on each pre- and post-registered image set were labeled by one blinded radiologist who had 5 years’ experience. Euclidean displacement was calculated according to the coordinates of the anatomic landmarks and the mean distance was used to evaluate the improvement in displacement after registration. And the Euclidean displacement was compared among the directly subtraction, commercial vendor method subtraction(Inline Liver Registration, Siemens Healthcare) and our method.Results
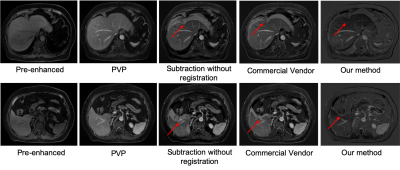
In comparison with manually annotated ground truth data, our model achieved an average DICE coefficient of 0.8624.The mean displacements of the anatomic landmarks are listed in Table 2. The average Euclidian displacement was significantly lower after registration compared to the non-registration ones. Further, our deeply supervised 3D U-net based method performed better than the MRI vendor commercial method. A representative case was shown in Figure 3. The average processing time for automatic liver subtraction was 0.88 ± 0.14 seconds for each data set on a personal computer with 64GB of system memory and a graphics processing unit with 11 GB of video memory (2080 Ti, NVIDIA, Santa Clara, CA, USA).Discussion
Accurate subtraction of the unenhanced image removes pre-existing T1 hyperintensity, which increasing the lesion contrast compared with the liver background. Registration of hepatic motion is of clinical interest in multiphase breath-hold DCE MR examinations to reduce the subtraction artifacts and improve lesion characterization. Our results demonstrate that the proposed method can improve the registration performance of pre- and post-contrast images through the mean displacements of the anatomic landmarks. This study has several limitations. First, the sample size was small for achieving a large-scale validation of the robustness of the subtraction and registration algorithm. Second, more quantitative indicators, such as temporal intensity smoothness, lesion volume change, and RMSE of TICs will be used to evaluate the registration performance in the future studies. Third, the subjective analysis of the subtraction results should be involved.Conclusion
We developed a deeply supervised 3D U-net based registration algorithm and automatically generate the subtraction liver images in DCE-MRI.Acknowledgements
NoReferences
[1] Yu JS, Kim YH, Rofsky NM. Dynamic subtraction magnetic resonance imaging of cirrhotic liver: assessment of high signal intensity lesions on nonenhanced T1-weighted images. J Comput Assist Tomogr 2005; 29:51–58
[2] Choi S H, Kim S Y, Lee S S, et al. Subtraction Images of Gadoxetic Acid–Enhanced MRI: Effect on the Diagnostic Performance for Focal Hepatic Lesions in Patients at Risk for Hepatocellular Carcinoma[J]. Ajr Am J Roentgenol, 2017:1-8.
[3] Sun Y, Zhu Q, Huang M, et al.Liver DCE-MRI registration based on sparse recovery of contrast agent curves. Med Phys. 2021 Aug 27.
[4] Hasenstab KA, Cunha GM, Higaki A et al.Fully automated convolutional neural network-based affine algorithm improves liver registration and lesion co-localization on hepatobiliary phase T1-weighted MR images. Eur Radiol Exp, 2019, 3(1):43.
[5] Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T., Ronneberger, O.: 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 424–432. Springer, Cham.
Figures