4179
Deep learning for term neonate hypoxic ischemic encephalopathy diagnosis on structural brain MR images: a retrospective study1Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China, 2Department of Artificial Intelligence, Julei Technology Company, Wuhan, China
Synopsis
This study aims to diagnose objectively term neonate hypoxic ischemic encephalopathy (HIE) by using deep learning network to extract deep information from multiple modalities of conventional magnetic resonance (MR) images. Neonate HIE diagnosis accuracy is restricted to lesion diversity, MR images quality, high interobserver variability. The network got high diagnosis accuracy in the ROC curve. The network could detect severe neonate HIE with characteristic appearance such as basal ganglia injury and periventricular leukomalacia. The network can help diagnosis neonate HIE objectively without the effect of different radiologist experience and contribute to risk stratification and clinical decision making.
introduction and purpose
Hypoxic ischemic encephalopathy is caused by a prolonged hypoxic event or diffuse brain blood flow reduction in the perinatal period with high disability and mortality. Structural MR imaging is standard tool to detect the location, distribution and severity[1]. Because of complicated patterns, different lesions of the injury and inconsistent MR imaging scan efficacy on different scanners, high interobserver variability influenced the diagnosis and therapeutic schedule seriously. We aimed to develop a deep learning network for hypoxic ischemic encephalopathy diagnosis by using structural MR imaging. Objective deep learning for high consistency in diagnosis contributes to precise clinical decision and risk stratification[2].Materials and methods
Patients: With the approval of the institutional review board, the study cohort comprised 645 term neonates (median gestational age 38.55+2.59weeks) including 303 patients of hypoxic ischemic encephalopathy and 342 healthy controls. Fulfillment of the criteria for the diagnosis of hypoxic ischemic encephalopathy served as ground truth.Imaging: on several MRI scanners (Achieva 1.5T, Philips, Amsterdam, Netherlands; Discovery 750, GE Health Care, Waukesha, Wisconsin, USA; United Imaging 780, Shanghai, China;)
Data processing: An end-to-end 3D ResNet34 deep learning architecture served as the diagnosis model, respectively using axial T1-weighted images and T2-weighted images as input to detect the presence of hypoxic ischemic encephalopathy. All images were preprocessed with an MRI automatic processing pipeline. The preprocessing steps included bias correction, intensity normalization, and anisotropic diffusion filtering. First, 3D ResNet34 networks with u-shape framework was trained for brain tumor segmentation in BraTS2017 using T1 and T2 modal, respectively. And the pre-trained 3D ResNet34 networks was fine-tuned in the train set with smaller initial learning rate. Finally, the two trained 3D ResNet34 networks were formulated ensemble model and evaluated in test set.
Results
In the test set, the accuracy in combination of T1WI and T2WI achieved 81.4%, superior to single T1WI or T2WI (79.8%, 76.7%, respectively). Besides, the model reached an AUC of 0.89 for the combination of T1WI and T2WI, higher than single T1WI and T2WI (0.86, p<0.05). The sensitivity and specificity for single T1WI and T2WI was 82.0%, 77.9% and 82.0%, 72.1%, respectively, which was improved in combination of T1WI and T2WI (85.2%, 77.9%)Discussion and conclusion
The deep learning network achieved high performance in HIE diagnosis for multimodal structural MRI (T1WI+T2WI), which outperformed T1WI or T2WI alone. These findings suggested multimodal MRI provide more features than single modal for diagnosis in neonate HIE, which was extracted by the network effectively. As for single MRI modal, T1WI showed higher diagnosis accuracy than T2WI in the deep learning network. That indicated that the network was more sensitive to hyperintense lesion such as basal ganglia injury in T1WI. The network could detect severe neonate HIE with characteristic appearance such as basal ganglia injury and periventricular leukomalacia, contributing to screening severe HIE for early medical intervention. Early diagnosis using objective methods is quite important for parental decision-making and neonatal management. To sum up, this study validates deep learning based radiomics for diagnosis in neonate HIE, suggesting deep learning based radiomics feature markers can be used to improve interobserver variability and diagnosis accuracy.Acknowledgements
Funding: This project was supported by National Natural Science Foundation of China (No.81730049)References
[1] TRIVEDI S B, VESOULIS Z A, RAO R, et al. A validated clinical MRI injury scoring system in neonatal hypoxic-ischemic encephalopathy [J]. Pediatr Radiol, 2017, 47(11): 1491-9.
[2] WEISS R J, BATES S V, SONG Y, et al. Mining multi-site clinical data to develop machine learning MRI biomarkers: application to neonatal hypoxic ischemic encephalopathy [J]. J Transl Med, 2019, 17(1): 385.
Figures
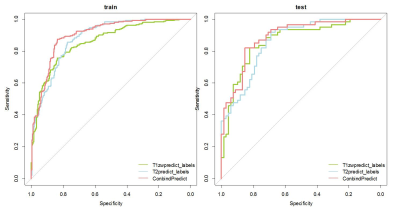
FIG1: Receiver operating characteristic (ROC) for neonate hypoxic ischemic encephalopathy diagnosis in the train set and test set.
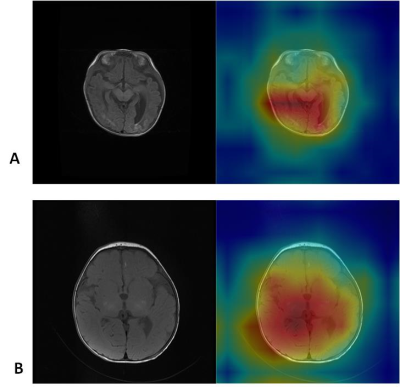
FIG2 :CAMs (class activation maps) depicting the lesion regions of the image that the network preferentially emphasizes when diagnosing neonate HIE on individual scan slices. The upper row shows the T1WI slice with characteristic lesion in original image (upper left) and CAM overlay of the most confident prediction of the model (upper right). The lower row shows basal ganglia injury in the same way.