4172
Non-contrast-enhanced mDixon water-fat separation whole-heart coronary MRA with a deep learning constrained Compressed SENSE reconstruction1Department of Radiology, The First Hospital of Jilin University, Changchun, China, 2Department of Cardiovascular center, The First Hospital of Jilin University, Changchun, China, 3Philips Healthcare, Beijing, China
Synopsis
The conventional 3D whole-heart free-breathing coronary MR angiography suffers from a long scan time. However, using very high acceleration factors leads to degradation of image quality due to insufficient noise removal. In this study, we use Compressed-SENSE Artificial Intelligence (CS-AI) framework to acquire highly accelerated 3D non-contrast-enhanced mDixon water-fat separation whole-heart CMRA. The result shows that CS-AI reconstruction can significantly decrease scan time with sufficient image quality compared to Compressed-SENSE(CS) and might be clinically useful in assessment of coronary artery disease.
Introduction
Three dimensional (3D) whole-heart free-breathing coronary MR angiography (CMRA) is a non-invasive diagnostic approach without radiation and has shown potential for the diagnosis of coronary artery disease (CAD). MRI chemical shift-based water-fat separation methods such as mDixon can provide excellent water and fat separation and can be used for CMRA1,2. However, CMRA still requires very long scan times due to the need of dealing with respiratory motion. Combined sensitivity encoding (SENSE) and compressed sensing (Compressed SENSE, CS) achieves scan time reduction beyond that possible with conventional parallel imaging acceleration. However, using very high acceleration factors leads to degradation of image quality due to insufficient noise removal3,4. More recently, deep learning-based algorithms have been combined with CS to overcome these challenges by learning optimal reconstruction parameters from the data itself and showed superior performance 5-8.Purpose
The purpose of this study was to acquire highly accelerated 3D non-contrast-enhanced mDixon water-fat separation whole-heart CMRA using the Compressed-SENSE Artificial Intelligence (CS-AI) framework and to compare impact of different acceleration factors(AF) on the image quality of conventional CS and CS-AI.Methods
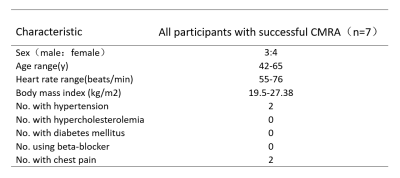
This study was approved by our institutional Ethics Committee and written informed consent was obtained from all subjects. A total of 7 participants were successfully examined on a 3.0T system (Ingenia Elition, Philips Healthcare), including 3 men and 4 women(mean age 53.43years, range: 42-65 years). Four participants were volunteers without known cardiovascular disease and the others were patients with suspected CAD. The 3D mDixon sequences were acquired with four different AF(4.5,6,8,10) and reconstruction by CS (CS 4.5,CS 6,CS 8,CS 10) and CS-AI (CS-AI 4.5,CS-AI 6,CS-AI 8,CS-AI 10). Following parameters were common to all examinations : TR/ TE1/TE2 = 4.4/1.42/2.6 ms; flip angle = 10; FOV = 265 X301 X 112 mm; ACQ matrix M×P = 176×201; ACQ voxel MPS (mm)= 1.51 X 1.50 X 1.50;REC voxel MPS (mm) = 0.75 X 0.75 X 0.75; slice thickness (mm)/gap = 1.5/-0.75;gating window:5.All the scans were exported to a workstation with image reconstruction software(IntelliSpacePortal, Version 10.1, Philips). Quantitative image analysis was performed by a radiologist with more than 8 years of experience. Signal to noise ratio (SNR) and contrast to noise ratio (CNR) were calculated by setting the coronary arteries(mainly right coronary artery) as the region of interest (ROI) and setting the left ventricular myocardium as a contrast. The image quality of CS and CS-AI was visually assessed by two experienced radiologists according to the 18-segment NASCI classification and a four-point-scale9,10. The image quality score over 3 was thought to be good enough for diagnosis. The Friedman test and Dunn’s host-pot analysis was performed to test for an influence of the different sequences. P values< 0.05 was considered significant.Results and Discussion
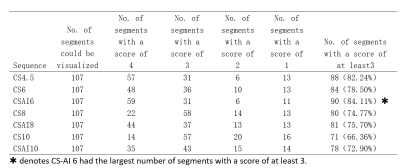
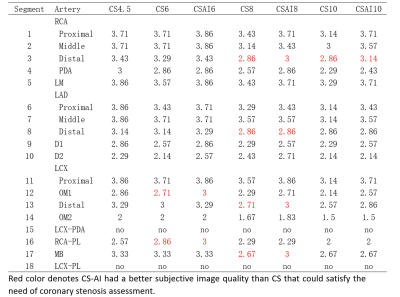
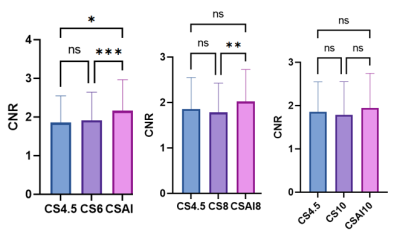
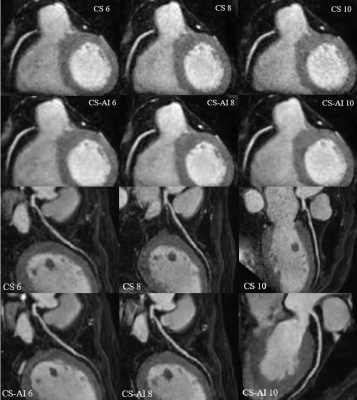
The study participants’ characteristics are summarized in Table1. Scan time decreased with increasing acceleration factors (mean scan time are as follows, CS4.5:9min25s, CS6/CS-AI6:6min30s, CS8/CS-AI8: 5min2s, CS10/CS-AI10: 4min3s). For quantitative analysis of image quality, The CNR values of CS6,CS-AI6 were significantly higher than CS 4.5(P<0.05),and the CS-AI6 sequence was rated highest for CNR value. The CNR values between CS8 and CS-AI8 were significantly different(P<0.01) ,but there were no statistical difference between the CNR values of CS 10,CS-AI 10 and CS4.5(Figure1). All of the SNR values of CS4.5, CS6, CS-AI6, CS8, CS-AI8, CS10, CS-AI10 had no significantly difference. For subjective analysis, CS-AI 6 had the largest number of segments with a score of at least 3, following by CS-AI 8 (Table2) . All of the segments of RCA, LAD, LCX had mean image quality scores larger than 3 in CS6 and CS-AI6; in CS 8 suquence, the mean image quality scores of RCA distal, LAD distal, LCX distal were smaller than 3, but in CS-AI8 the corresponding segments had a better subjective image quality with mean scores of 3, 2.86 and 3, respectively. All the subject scores of CS-AI sequences were significantly higher than time-equivalent CS(all P<0.05).The mean image quality scores for all segments are listed in the Table3. Representative 3D whole heart mDixon CMRA images using the CS and CS-AI reconstructions with different AF are shown in Figure 2.Conclusion
The results of our preliminary study suggest that the application CS-AI reconstruction can significantly decrease scan time in 3D non-contrast-enhanced whole heart mDixon water-fat separation CMRA, with better image quality compared to conventional CS with same acceleration factors. CS-AI6 provids optimal image quality with obvious scan time reduction(30.97%); CS-AI8 provids acceptable image quality with significant scan time reduction(46.55%); Although further clinical investigation is needed, 3D mDixon water-fat separation CMRA with CS-AI reconstruction might be clinically useful, especially for “one-stop-shop” cardiac MRI , in assessment of coronary artery disease.Acknowledgements
No acknowledgement found.References
1. Hongfei Lu, Jiajun Guo, Hang Jin, et al. Assessment of Non-contrast-enhanced Dixon Water-fat Separation Compressed Sensing Whole-heart Coronary MR Angiography at 3.0 T: A Single-center Experience. Acad Radiol.2021;05.009:1-9.
2. Hongfei Lu, Shihai Zhao, Hang Jin, et al. Clinical Applicationof Non-Contrast-Enhanced Dixon Water-Fat Separation Compressed SENSE Whole-Heart Coronary MR Angiography at 3.0 T With and Without Nitroglycerin. J. MAGN. RESON. IMAGING. 2021; DOI10.1002/jmri.27829.
3. Gao T, Lu Z, Wang F, Zhao H, Wang J, Pan S. Using the Compressed Sensing Technique for Lumbar Vertebrae Imaging: Comparison with Conventional Parallel Imaging. Curr Med Imaging 2021; 17(8): 1010-7.
4. Iuga AI, Abdullayev N, Weiss K, et al. Accelerated MRI of the knee. Quality and efficiency of compressed sensing. Eur J Radiol 2020; 132: 109273.
5. Knoll F, Zbontar J, Sriram A, et al. fastMRI: A Publicly Available Raw k-Space and DICOM Dataset of Knee Images for Accelerated MR Image Reconstruction Using Machine Learning. Radiol Artif Intell. 2020;2(1):e190007.
6. Knoll F, Murrell T, Sriram A, et al. Advancing machine learning for MR image reconstruction with an open competition: Overview of the 2019 fastMRI challenge. Magn Reson Med. 2020;(January):mrm.28338.
7. Pezzotti N, Yousefi S, Elmahdy MS, et al. An Adaptive Intelligence Algorithm for Undersampled Knee MRI Reconstruction. IEEE Access. 2020;8:204825-204838.
8. Pezzotti N, de Weerdt E,Yousefi S, et al. Adaptive-CS-Net: FastMRI with Adaptive Intelligence. arxiv. 2019;(NeurIPS).
9. Abbara S, Blanke P, Maroules CD, et al. SCCT guidelines for the performance and acquisition of coronary computed tomographic angiography: a report of the society of Cardiovascular Computed Tomography Guidelines Committee: Endorsed by the North American Society for Cardiovascular Imaging (NASCI) [J]. J Cardiovasc Comput Tomogr, 2016, 10(6): 435‑449. DOI: 10.1016/j.jcct.2016.10.002.
10. He Y, Pang JN, Dai Q, et al. Diagnostic Performance of Self-navigated Whole Heart Contrast-enhanced Coronary 3-T MR Angiography. Radiology2016; 281(2):401–408.
Figures