4123
Convolutional neural networks to differentiate hypertrophic cardiomyopathy from hypertensive heart disease based on cardiac cine imaging1Department of Cardiology, Ren Ji Hospital,Shanghai Jiao Tong University School of Medicine, Shanghai, China, 2Institute of Medical Imaging Technology, School of Biomedical Engineering, Shanghai Jiao Tong University, shanghai, China
Synopsis
Differentiating hypertrophic cardiomyopathy (HCM) from hypertensive heart disease (HHD) is important yet challenging. In this study, we compared 9 convolutional neural network (CNN) models based on cardiac MR cine imaging only for differentiation of the two diseases. We show that the dynamic information contained in cine about myocardial contraction and relaxation is crucial for accurate differentiation. By leveraging this information, we achieved a testing accuracy of 86.8% ± 3.5% in a cohort including 190 HCM and 113 HHD subjects. The results show that cine-based CNN is reasonably accurate for differentiation of HCM and HHD.
introduction
Hypertrophic Cardiomyopathy(HCM) and Hypertensive Heart Disease(HHD) are two different diseases with similar morphological features. Differentiating HCM from HHD is of significant clinical value, yet an accurate differentiation bears great challenges, especially for patients with both HCM indications and history of hypertension. In this study, we sought to develop a deep neural network to differentiate HCM from HHD based on cardiac magnetic resonance(CMR) cine imaging. In particular, we hypothesize that dynamic information contained in cine imaging is important for accurate classification of the two diseases by neural networks.Methods
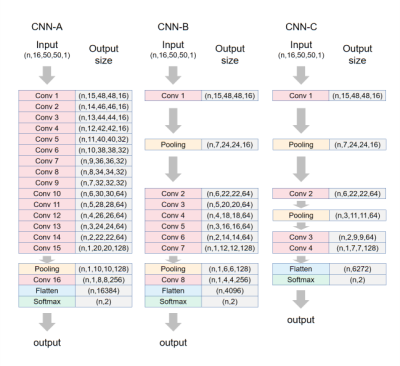
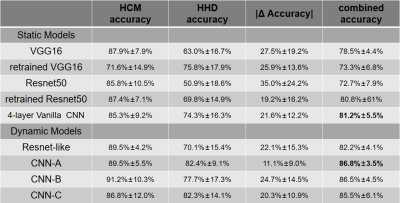
A total of 342 HCM and HHD patients who underwent CMR exam was included in this study based on diagnosis. CMR images were acquired by experienced radiologists using a 3T MRI scanner (Ingenia, Philips) equipped with a 16-channel spine coil and 16-channel torso coil. bSSFP cine imaging was performed in short-axis view with breath-holding. Typical cine parameters were: TR/TE/flip angle/slice thickness/bandwidth/resolution/number of phases/SENSE factor = 2.7ms/1.4ms/45°/5-10mm/1900(Hz/pixel)/1.6-2mm/30/2. All images were then cropped and interpolated to 100Í100 voxels with 1.5mm isotropic resolution. Seven central slices continuously selected from the stack of cines and 16 continuous phases (Phase 1 to 16 covering systole and early diastole) were used in classification. Nine deep neural network models were trained and tested. To verify the hypothesis about cruciality of dynamic information, we divided the 9 models into static models and dynamic models, where the former generated predictions for each end-diastolic 2D image and the latter for each movie. For static models, we included pre-trained VGG16 and Resnet50 networks, which were fine-tuned by freezing the first 4 blocks and training the remaining blocks1,2, completely-retrained VGG16 and Resnet50, and a 4-layer Vanilla CNN for comparison. For the dynamic models, we included a Resnet-like model, a 16-layer Vanilla CNN (CNN-A, Figure 1), and two simplified CNNs (CNN-B and CNN-C) respectively modified from CNN-A by replacing 8 convolutional layers with 1 pooling layer and 12 convolutional layers with 2 pooling layers. Training was performed with data augmentation including rotation (±15°) and translation(±10%), 10-fold cross validation, and with the Adam algorithm (learning rate=10-5), and batch sizes of 32-128. Each model generated a probability score for each of the 7 slices in each patient. HCM was predicted for the patient if the average score over the 7 slices was greater than 0.5. In addition to relying solely on neural networks, we also leveraged the concept of ensemble learning by combining the prediction result of each neural network model with left ventricle ejection fraction (LVEF) to generate a prediction for each patient. Three binary classification models, including logistic regression(LR), support vector machine(SVM), and K-nearest neighbor (KNN) were compared.3Results
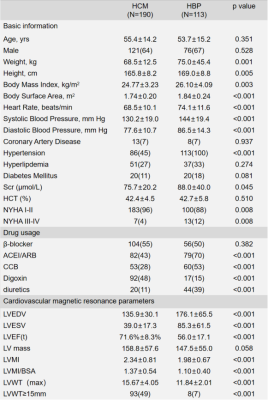
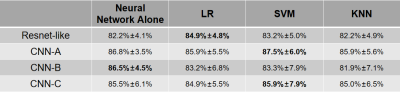
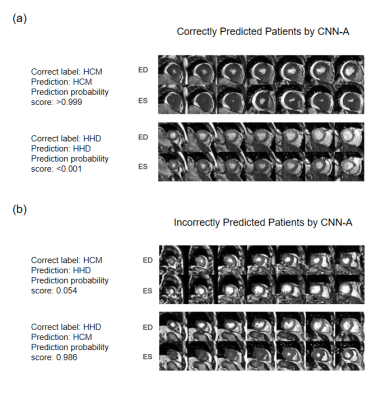
After excluding patients with other documented heart diseases, 303 patients including 190 HCM and 113 HHD were included for further analysis. Detailed patient characteristics information is shown in Table 1. Note that although the HCM group had a significantly lower hypertension prevalence, 45% of HCM subjects still had hypertension(p<0.001).Table 2 shows the classification accuracy of the 9 neural network models. The metric |Δ Accuracy| refers to the absolute value of HCM accuracy - HHD accuracy, indicating classification bias mostly caused by group size imbalance. Among all static models, surprisingly, the simplest 4-layer CNN static model reached the highest combined accuracy(81.2% ± 5.5%) and second-lowest |Δ Accuracy|(21.8% ± 12.2%).4 The performance of dynamic models was overall better than static models, suggesting dynamic information is important for accurate prediction. Within the dynamic models, CNN-A had the best diagnostic accuracy (86.8% ± 3.5%) and lowest |Δ Accuracy|(11.1%±9.0%).Table 3 shows the result of ensemble learning. The SVM method achieved better accuracy than the other two approaches. However, compared with the result obtained by networks alone, there was only a small change of accuracy (87.5% vs. 86.8%) accompanied by a higher standard deviation (6.0% vs. 3.5%).Figure 2 shows four patients, two correctly predicted and the other two incorrectly predicted by CNN-A. The correctly predicted patients had rather typical morphological and functional features, including asymmetric hypertrophy and apical obliteration for the HCM patient and symmetric hypertrophy for the HHD patient.5 The incorrectly predicted patients, however, showed a morphology quite opposite to general impression, with the HCM patient showing a symmetric hypertrophy and HHD an asymmetric hypertrophy.Discussion and conclusions
The results showed that dynamic models performed better than the static models, suggesting that features about the left ventricular contraction and relaxation are important for accurate differentiation of the two diseases. A recent study including 53 HHD subjects and 108 HCM subjects performed by Ulf Neisius et.al showed 80% accuracy with use of radiomics.6 Our testing data accuracy is comparable and mildly higher than the previous study, potentially due to the use of deep neural networks and dynamic information. In conclusion, our result shows that convolutional neural networks based on MR cine dataset only are feasible and reasonably accurate for differentiation of HCM vs HHD.Acknowledgements
No acknowledgement found.References
1. Simonyan, K., & Zisserman, A. (2014). Very deep convolutional networks for large-scale image recognition. CoRR, abs/1409.1556.
2. Kaiming He, Xiangyu Zhang, Shaoqing Ren, and Jian Sun. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pages 770–778, 2016.
3. Scikit-learn: Machine Learning in Python, Pedregosa et al., JMLR 12, pp. 2825-2830, 2011.
4. Morid MA, Borjali A, Del Fiol G. A scoping review of transfer learning research on medical image analysis using ImageNet. Comput Biol Med. 2021 Jan;128:104115. doi: 10.1016/j.compbiomed.2020.104115. Epub 2020 Nov 13. PMID: 33227578.
5. Rapezzi C, Quarta CC, Obici L, Perfetto F, Longhi S, Salvi F, Biagini E, Lorenzini M, Grigioni F, Leone O, Cappelli F, Palladini G, Rimessi P, Ferlini A, Arpesella G, Pinna AD, Merlini G, Perlini S. Disease profile and differential diagnosis of hereditary transthyretin-related amyloidosis with exclusively cardiac phenotype: an Italian perspective. Eur Heart J. 2013 Feb;34(7):520-8. doi: 10.1093/eurheartj/ehs123. Epub 2012 Jun 28. PMID: 22745357.
6. Neisius U, El-Rewaidy H, Nakamori S, Rodriguez J, Manning WJ, Nezafat R. Radiomic Analysis of Myocardial Native T1 Imaging Discriminates Between Hypertensive Heart Disease and Hypertrophic Cardiomyopathy. JACC Cardiovasc Imaging. 2019 Oct;12(10):1946-1954. doi: 10.1016/j.jcmg.2018.11.024. Epub 2019 Jan 16. PMID: 30660549; PMCID: PMC7032053.
Figures