4083
NiBabies: A robust preprocessing workflow tailored for neonate and infant MRI1Stanford University, Stanford, CA, United States, 2University of North Carolina School of Medicine, Chapel Hill, NC, United States, 3Oregon Health & Science University, Portland, OR, United States, 4University of Minnesota, Minneapolis, MN, United States, 5Washington University in St. Louis, St. Louis, MO, United States, 6Harvard Medical School, Boston, MA, United States, 7University of Lausanne, Lausanne, Switzerland
Synopsis
Advances in both data acquisition and processing methods have given magnetic resonance imaging researchers (MRI) a plethora of options on how best to clean and standardize data before statistical analysis. Recently, there has been a surge in standardized data processing workflows, but special populations, such infants, require modified techniques not normally found in general pipelines. Here we introduce NiBabies, a robust and open-source structural and functional MRI preprocessing pipeline designed for infant populations.
Introduction
Magnetic resonance imaging (MRI) requires a set of preprocessing steps before any statistical analysis. In an effort to standardize preprocessing, we developed fMRIPrep (a preprocessing tool for functional MRI, fMRI, Esteban 2019a), and generalized its standardization approach to other neuroimaging modalities (NiPreps, Esteban 2019b). NiPreps brings standardization and ease of use to the researcher, and effectively limits the methodological variability (Botvinik-Nezer 2020) within preprocessing. fMRIPrep is designed to be used across wide ranges of populations; however it is designed for (and evaluated with) human adult datasets. Infant MRI (i.e., 0-2 years) presents unique challenges due to head size (e.g., reduced SNR and increased partial voluming (Mostapha 2019)) and rapid shifting in tissue contrast due to myelination. These and other challenges require a more specialized workflow. Here, we propose NiBabies, an open-source pipeline extending from fMRIPrep for infant structural and functional MRI preprocessing.Methods
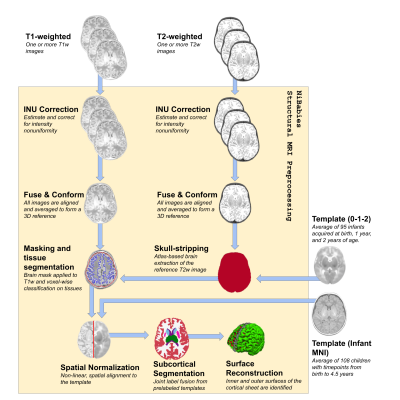
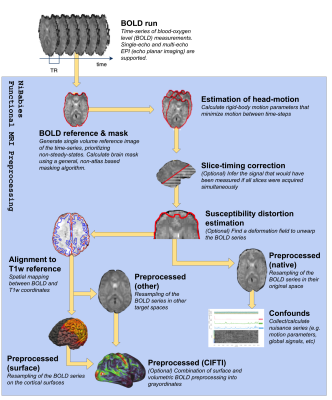
The NiBabies pipeline (Fig 1, Fig 2) is built on the developmental design laid out by fMRIPrep (Esteban 2019a), utilizing a symphony of tools from widely-available software packages with the goal of providing reliable preprocessing that adapts to diverse and idiosyncratic datasets. NiBabies leverages BIDS (Gorgolewski 2016) to adapt the workflow to the input data and to organize the preprocessed outputs (BIDS-Derivatives). NiBabies is developed following the principles and driven by the roadmap set by the NiPreps community, which establishes some coding standards, validation strategies and recommendations, transparent and thorough reporting, and reproducible environments with container technologies.Results
To address some of the fundamental differences between adult and infant MRI, NiBabies adapts fMRIPrep’s workflow in a number of ways:Refinement of brain extraction - Similar to fMRIPrep, NiBabies uses an atlas-based registration method for anatomical brain extraction. However unlike fMRIPrep, the infant’s T2-weighted image is used for normalization to the target atlas, to leverage the more tissue-specific T2 contrast. The pipeline defaults to using the infant 0-1-2 atlas (Shi 2011), which provides three atlases with distinct points in infant development. The proper time point is automatically assigned based on age and can be overridden by the user, which is made possible by TemplateFlow (Ciric 2021).
Support for multi-atlas segmentation - The sheer difference in size and contrasts between adult and infant brain structures requires increased attention to produce accurate cortical and subcortical segmentation. NiBabies allows users to provide one or more pre-labeled images to be used for label fusion. Each labeled image provides a voxelwise weighted vote towards the final segmentation, increasing the robustness by considering diverse brain morphologies (Wang 2013).
Improvement of surface reconstruction - NiBabies integrates Infant FreeSurfer’s surface extraction, which accounts for the nuances of the developing infant brain. A significant deviation from the adult FreeSurfer reconstruction pipeline is the cortical modeling shift in weight from intensity contrast of the images towards the volumetric segmentations; this shift was found to improve surface fitting performance in infants (Zöllei 2020).
Fine-grain subcortical alignment - NiBabies can produce CIFTI-2 files, combining surface and volumetric BOLD data into a single file. These files follow the “grayordinates” specification proposed by the Human Connectome Project (Glasser 2013), and require resampling the volumetric data to an MNI template generated from adult brains, which contains subcortical structures drastically different in size and shape compared to infants. NiBabies ensures each structure is independently registered to the template to improve overall accuracy.
Conclusion
NiBabies provides a transparent, modular workflow for minimal pre-processing, resulting in commonly desired derivatives. It provides maximum flexibility by producing analysis-agnostic results, thereby supporting (virtually) any strategy to downstream, higher-level analysis. Its open-source based modular design is also well suited for community contributions and improvements as the field advances. One such improvement will be replacing computation-intensive steps with convolution deep learning, as networks become increasingly efficient and accurate.Acknowledgements
This work has been supported by the NIMH (RF1MH121867; RAP, OE). OE receives support from the SNSF Ambizione Project “Uncovering The Interplay Of Structure, Function, and Dynamics of Brain Connectivity using MRI” (grant number PZ00P2_185872).References
Botvinik-Nezer R, Holzmeister F, Camerer CF et al. Variability in the analysis of a single neuroimaging dataset by many teams. Nature 582, 84–88 (2020). https://doi.org/10.1038/s41586-020-2314-9
Ciric R, Lorenz R, Thompson W, Goncalves M, MacNicol E, Markiewicz C, Halchenko Y, Ghosh S, Gorgolewski K, Poldrack R, Esteban O. 2021. TemplateFlow: a community archive of imaging templates and atlases for improved consistency in neuroimaging. https://doi.org/10.1101/2021.02.10.430678
Esteban O, Markiewicz C, Blair RW, Moodie C, Isik AI, Aliaga AE, Kent J, Goncalves M, DuPre E, Snyder M, Oya H, Ghosh S, Wright J, Durnez J, Poldrack R, Gorgolewski KJ. (2019a). fMRIPrep: a robust preprocessing pipeline for functional MRI. Nat. Methods 16, 111-116, https://doi.org/10.1038/s41592-018-0235-4
Esteban, O, Wright, J, Markiewicz, CJ, Thompson, WH, Goncalves, M, Ciric, R, … Poldrack, R. (2019b). NiPreps: enabling the division of labor in neuroimaging beyond fMRIPrep. https://doi.org/10.31219/osf.io/ujxp6
Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, Xu J, Jbabdi S, Webster, M, Polimeni JR, Van Essen DC, Jenkinson M. 2013. The minimal preprocessing pipelines for the Human Connectome Project. NeuroImage 80, 105-124, https://doi.org/10.1016/j.neuroimage.2013.04.127
Gorgolewski KJ, Auer T, Calhoun VD, Craddock RC, Das S, Duff EP, Flandin G, Ghosh SS, Glatard T, Halchenko YO, Handwerker DA, Hanke M, Keator D, Li X, Michael Z, Maumet C, Nichols BN, Nichols TE, Pellman J, Poline JB, Rokem A, Schaefer G, Sochat V, Triplett W, Turner JA, Varoquaux G, Poldrack RA. 2016. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci. Data 3, 160044. https://doi.org/10.1038/sdata.2016.44
Mostapha M, Styner M. 2019. Role of Deep Learning in Infant Brain MRI Analysis. Magn Reson Imaging 64, 171-189, https://doi.org/10.1016/j.mri.2019.06.009
Shi F, Yap PT, Wu G, Jia H, Gilmore JH, Lin W, Shen D. 2011. Infant Brain Atlases from Neonates to 1- and 2-Year-Olds. PLOS ONE 6(4): e18746. https://doi.org/10.1371/journal.pone.0018746
Wang H, Yushkevich PA. Groupwise segmentation with multi-atlas joint label fusion. Med Image Comput Comput Assist Interv. 2013;16(Pt 1):711-718. https://doi.org/10.1007/978-3-642-40811-3_89
Zöllei L, Iglesias JE, Ou Y, Grant PE, Fischl B. Infant FreeSurfer: An automated segmentation and surface extraction pipeline for T1-weighted neuroimaging data of infants 0-2 years. Neuroimage. 2020 Sep;218:116946. https://doi.org/10.1016/j.neuroimage.2020.116946.