4058
Enhanced detection of paramagnetic fluorine-19 MRI agents using compressed sensing zero echo time sequence1Department of Electrical and Computer Engineering, University of California San Diego, La Jolla, CA, United States, 2Department of Radiology, University of California San Diego, La Jolla, CA, United States
Synopsis
To accelerate the imaging speed and improve the signal-to-noise ratio of three-dimensional, whole body 19F MRI, we describe an ultrafast compressed sensing data acquisition scheme adapted for a zero echo time (ZTE) pulse sequence, with data reconstruction via a sparsity-promoting algorithm using a nonuniform fast Fourier transform (NUFFT) sensing operator. These methods are well suited for MRI of labeled cells using 19F tracer agents, particularly paramagnetic metallo-perfluorocarbon nanoemulsions. In phantom data and an in vivo immunotherapy mouse model, we show the feasibility of boosting MRI detection sensitivity by greater than an order of magnitude without increasing total scan time.
Introduction
Fluorine-19 MRI is an emerging technique offering specific detection of labeled cells in vivo. Lengthy acquisition times and modest signal-to-noise ratio (SNR) makes 3D spin-density weighted 19F imaging challenging. Recent advances in tracer paramagnetic metallo-perfluorocarbon (MPFC) nanoemulsion probes have shown multifold SNR improvements due to an accelerated 19F T1 relaxation rate and a commensurate gain in imaging speed. However, T2-reduction and increased linewidth limit the amount of metal additive in MPFC probes, thus constraining the ultimate SNR. To overcome these barriers, we describe a compressed sampling (CS) scheme, implemented using a zero echo time (ZTE) sequence, with data reconstructed via a sparsity-promoting algorithm. Our results show that CS-ZTE acquisitions achieve at least an order of magnitude improvement in the limit of detection (LOD) for MPFC in phantoms; in vivo, CS-ZTE MRI data display comparable detection benefits, with enhanced detection of sparse cell densities in dendritic cell (DC) grafts in mice.Methods
The 3D radial k-space sampling pattern employed by ZTE pulse sequence (Fig. 1A) is constructed from projections with endpoints on a spherical surface. To accelerate k-space acquisitions, we undersample in angular directions of the radial trajectory by drawing random points on a 3D spherical surface1. These surface endpoints, associated with angular directions $$$(\theta, \Psi) $$$, were distributed in a Poisson disk and were then transformed into a table that controls the magnetic field gradients used for MRI acquisitions. We acquired k-space radial data with an undersampling factor, denoted $$$u$$$, varying from 1 (fully sampled) to 8 (Fig. 1B). Reconstruction of CS-ZTE data was divided into two steps, including standard algebraic preprocessing2 and sparsity-promoting regularized reconstruction. The second step involves modifying off-the-shelf sparse solvers3-4 with regularization parameters chosen using a Pareto frontier curve in a data-driven fashion.To validate the performance of CS-ZTE, MRI experiments were performed on an 11.7 T MRI scanner with a 40 mm dual-tuned 1H/19F birdcage resonator. Initial 19F phantoms were comprised of three embedded tubes containing dilutions (100%, 50%, 25%) of MPFC nanoemulsion5 with 19F T1 and T2* values equal to 7 ms and 1.5 ms, respectively. Phantom acquisition parameters were 60 kHz bandwidth, 643 voxels, (30 mm)3 field of view (FOV), 10° tip angle, 1.6 ms TR and NA = 10. For the in vivo mouse study, PFC or MPFC nanoemulsion at 2.5 mg/mL was added to fetal skin-derived dendritic cells (FSDCs)6 in culture and co-incubated for four hours followed by a wash step. Labeled FSDCs were then injected into the left (PFC) and right (MPFC) footpad (5×106 cells per side) of female, 6-8 week, C57BL/6 mice ($$$N=3$$$). Three-dimensional 19F images were acquired using a fixed scan time of 10 min for conventional ZTE and CS-ZTE ($$$u=8$$$). The in vivo studies used the same imaging parameters as the phantom, apart from the FOV (60 mm)3 and the number of averages (NA = 30). Additionally, 1H anatomical images were acquired using a 2D RARE (factor of 8) with TE/TR = 23/2500 ms, FOV = (60mm)2, 3842 matrix, NA = 4, and 0.47 mm thick slices.
Results
For fixed scan time, significant SNR gains are observed in phantom experiments (Fig. 1C) using the sparsely sampled trajectory and CS reconstruction algorithm. For the MPFC data acquired with $$$u=8$$$, the measured LOD7 displays an order of magnitude enhancement, from ~$$$3\times 10^{16}$$$ to ~$$$3\times 10^{15}$$$. 19F atoms per voxel compared to the ZTE method ($$$u=1$$$, Fig. 1C). We show that the CS-ZTE method is effective for in vivo scans and yields substantial sensitivity improvements (Fig. 2). Fig. 2A shows representative in vivo 1H (grayscale) / 19F (pseudo-color) MRI results in mice ($$$N=3$$$) that were injected with PFC- and MPFC-labeled FSDCs in the left and right hind footpad, respectively. For all 3D acquisitions, the scan time was fixed at 10 min with (0.94mm)3 isotropic voxels. The CS-ZTE sequence readily detects the MPFC labeled cells, which appears as a hot-spot in the right foot pad (Fig. 2A). Cells labeled with metal-free PFC (left footpad) fall below the detection limits and cannot be observed by the optimized sequence. The corresponding image voxel histogram distributions of signals and noise are displayed in Fig. 2B. With the CS-ZTE method, we observe a ~7-fold increase in vivo in the mean 19F signal intensity using CS undersampling ($$$u=8$$$) (Fig. 2B). Importantly, the wider intensity range and larger number of signal-positive voxels observed with the CS-ZTE method highlights its ability to detect smaller numbers of cells per voxel. From the noise perspective, CS-ZTE displays a slight increase in estimated standard deviation (~1.1-fold) (Fig. 2B). This is less than the theoretical ~2.8-fold ($$$\sqrt{u}$$$, where $$$u=8$$$) increment due to the denoising effects of the sparsity-promoting reconstruction algorithm.Discussion and Conclusion
Overall, using CS-ZTE we observe detection enhancement from three distinct mechanisms, including additional signal averaging enabled by CS undersampling, the intrinsic denoising attributes of the sparsity-promoting reconstruction algorithm, as well as enhanced acquisition speed and a subsequent increase in signal averaging when MPFC materials are used. In conclusion, we show that the CS-ZTE is effective at significantly improving SNR and detection sensitivity for 3D 19F MRI. The CS-ZTE method displays tremendous potential for future preclinical therapeutic cellular-molecular studies.Acknowledgements
Funding for ETA was provided by National Institutes of Health (NIH) grants R01-EB024015, R01-CA139579 and the California Institute for Regenerative Medicine LA1-C12-06919.References
1. Johnson E, Pauly K, Pauly J. Spherical-surface Poisson disc point selection for radial-trajectory. In Proceedings of the 25th Annual Meeting of ISMRM; 2017; Honolulu, HI.
2. Weiger M, Pruessmann K. MRI with Zero Echo Time. Emagres. 2012;1(2):311-321.
3. Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine. 2007;58(6):1182-1195.
4. Fessler JA, Sutton BP. Nonuniform fast Fourier transforms using min-max interpolation. IEEE transactions on signal processing. 2003;51(2):560-74.
5. Jahromi AH, Wang C, Adams SR, et al. Fluorous-soluble metal chelate for sensitive fluorine-19 magnetic resonance imaging nanoemulsion probes. ACS nano. 2018;13(1):143-151.
6. Ahrens ET, Flores R, Xu H, et al. In vivo imaging platform for tracking immunotherapeutic cells. Nature biotechnology. 2005;23(8):983-987.
7. Ahrens ET, Helfer BM, O'Hanlon CF, Schirda C. Clinical cell therapy imaging using a perfluorocarbon tracer and fluorine‐19 MRI. Magnetic resonance in medicine. 2014;72(6):1696-701.
Figures
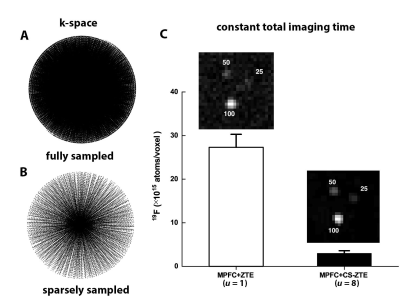
Figure 1: CS-ZTE undersampling patterns and corresponding acquisitions of MPFC 19F 3D MRI data of matrix size of 643, along with LOD evaluations. A. 3D radial fully sampled k-space trajectory (u=1, 6515 spokes). B. 3D radial undersampled k-space trajectory (u=8, 814 spokes). C. MPFC image acquired from CS-ZTE with the scan time of 3min. The values on slices represent percent dilution. The LOD (19F atoms/voxel) improvement from ~3×1016 to ~3×1015, represents an order of magnitude detection enhancement.
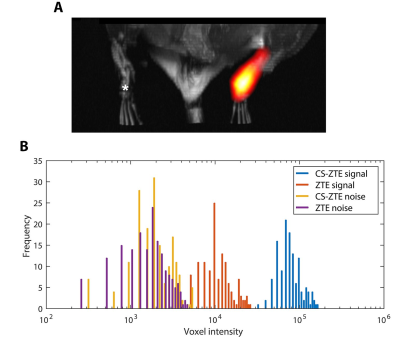
Figure 2: In vivo 3D 19F MRI mouse data showing PFC and MPFC labeled FSDCs. A. In vivo 1H (grayscale) / 19F (pseudo-color) MRI of mice displaying cell deposit ‘hot-spot’ in right footpad corresponding to the MPFC-labeled FSDCs. Cells labeled with iron-free PFC nanoemulsion (left footpad, asterisk) are below the LOD. B. Histogram of unthresholded 19F image data of CS-ZTE (u=8) and conventional ZTE. The 3D ROI, defined for both signal and noise regions, are kept constant across datasets.