4053
Multi-Task Accelerated MR Reconstruction Schemes for Jointly Training Multiple Contrasts1California Institute of Technology, Pasadena, CA, United States, 2Stanford University, Stanford, CA, United States
Synopsis
Model-based accelerated MRI reconstruction networks leverage large datasets to reconstruct diagnostic-quality images from undersampled k-space. To deal with inherent dataset variability, the current paradigm trains separate models for each dataset. This is a demanding process and cannot exploit information that may be shared amongst datasets. In response, we propose multi-task learning (MTL) schemes that jointly reconstruct multiple datasets. Introducing inductive biases to the network allows for positive information sharing. We test MTL architectures and weighted loss functions against single task learning (STL). Our results suggest that MTL can outperform STL across a range of dataset ratios for two knee contrasts.
Introduction
To reduce MRI scan time, various iterative reconstruction schemes have been investigated.1-4 Recently, deep learning approaches, which train a network to estimate the reconstructed image using retrospectively undersampled k-space, have shown superior efficacy over previous non-network based methods.5 However, these networks require sufficient collection of fully sampled k-space data from similar acquisition protocols as the test-time inference data.6 For example, to train multiple contrasts, the current paradigm is to collect multiple, fully sampled k-space data for each contrast and train each contrast-specific network separately to avoid domain shift.6-7 Considering the exceptionally large variability of MR images (i.e. different contrasts, orientations, anatomies, pulse sequences), separate training requires a large effort and limits the additional information that can be gained from multiple datasets.To address this barrier, we propose a novel multi-task learning (MTL) scheme that can jointly train a single network on a variety of datasets. MTL has recently gained traction in various areas,8–15 but has yet to be applied to MRI reconstruction. Our study investigates how this scheme can be useful for jointly training diverse datasets. The scheme jointly trains various fully sampled k-space datasets by treating them as different tasks within the same network. The network can train multiple tasks simultaneously and exploit shared, common features to prevent individual tasks from overfitting, thereby fostering better performance compared to conventional STL counterparts.
Methods
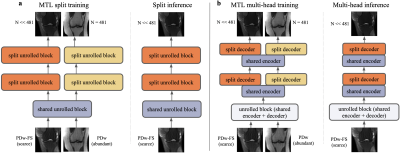
The baseline STL network is a typical unrolled network composed of a series of unrolled blocks.16 The MTL network structure is composed of shared layers and task-specific layers, each consisting of multiple unrolled blocks inside the layer. Two architectures are used in the study: split and multi-head (Figure 1). Both networks start with two shared blocks before splitting into ten task-specific blocks. In the split architecture, the task-specific blocks do not share further information (1a). In the multi-head architecture, the U-Net encoder continues to be shared amongst tasks, but the decoder is task-specific (1b).We consider three different loss weighting schemes. Naive weighting addresses the data imbalance in the loss function by weighting individual task losses in an inverse relation to dataset size. Uncertainty weighting9 treats the multi-task network as a probabilistic model and incorporates the homoscedastic uncertainty in the loss function. Dynamic weight averaging (DWA)14 assigns task losses based on the learning speed of each task.
Training and Inference
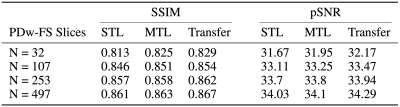
We use two public knee datasets16 that are available at mridata.org.17 The datasets contain 19 coronal proton density weighted (PDw) and 20 coronal proton density weighted fat suppression (PDw-FS) knee scans. In our experiments, PDw simulates the abundant dataset by using all 481 slices, and PDw-FS simulates the scarce dataset by using a percentage of the 497 slices.Our models are implemented in PyTorch and trained on NVIDIA Titan Xp GPUs with 12GB of memory. For the experiments, networks with 12 unrolled blocks are used to ensure convergence. To assess image quality, magnitude images are normalized between 0 and 1, and peak signal-to-noise ratio (pSNR), structural similarity index (SSIM), and normalized root mean square error (nRMSE) are used. During inference, k-spaces in the test-set are undersampled identically to guarantee fair comparisons.
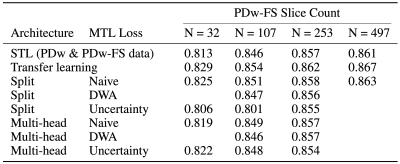
For this study, we mix and match the network architectures (split and multi-head) with the loss functions (naive, DWA, uncertainty) for a total of six MTL networks (Tables 2-3). MTL networks are jointly trained using 481 PDw slices and a percentage of PDw-FS slices. We also provide comparisons with transfer learning by taking the PDw baseline and fine-tuning all layers using PDw-FS data.6
Results
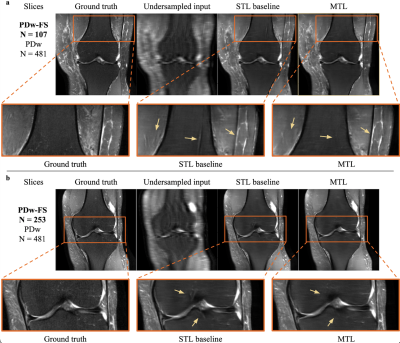
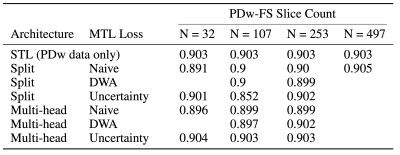
As seen in Table 1, an MTL network performs better than STL at every dataset ratio. At N = 107 and N = 253, the naive-weighted, split architecture dominates the other MTL architectures (Tables 2-3). Interestingly, the MTL network not only improves metrics for PDw-FS, but also for PDw at certain ratios (Tables 2-3). This suggests that there is positive information transfer from the scarce dataset to the abundant dataset as well.Qualitative examination also suggests that MTL reduces errors in reconstruction (Figure 2) at both ratios. A comparison between STL and MTL is seen in Figure 2 for two different inference slices. Interestingly, transfer learning performs better than MTL quantitatively (Tables 1-3).
Conclusion
Our framework introduces inductive biases in the network by enforcing the sharing of useful information between tasks. We see that MTL performs better than STL baselines across a range of abundant versus scarce ratios, for both PDw and PDw-FS datasets. Our finding that transfer learning marginally outperforms MTL suggests that MTL may be better suited for more dissimilar tasks such as different orientations, anatomies rather than multi-contrasts. Moreover, it is possible that once we use larger datasets, STL will dominate transfer learning, and MTL’s main competitor will be STL.Discussion
One noted difficulty is selecting an appropriate architecture and loss function, since negative transfer between the dataset can occur (Tables 2-3). All in all, our study provides a proof of concept that MTL can be successfully used in jointly training multiple contrasts for MRI reconstruction.Acknowledgements
This project is supported by NIH R01 EB009690 and NIH R01 EB029427, as well as GE Healthcare and the Marcella Bonsall Fellowship.References
1. Jong Chul Ye. Compressed sensing mri: a review from signal processing perspective. BMC Biomedical Engineering, 1, 03 2019. doi: 10.1186/s42490-019-0006-z.
2. Xinlin Zhang, Di Guo, Yiman Huang, Ying Chen, Liansheng Wang, Feng Huang, and Xiaobo Qu. Image reconstruction with low-rankness and self-consistency of k-space data in parallel mri, 2019.
3. Anagha Deshmane, Vikas Gulani, Mark Griswold, and Nicole Seiberlich. Parallel mr imaging. Journal of magnetic resonance imaging : JMRI, 36:55–72, 07 2012. doi: 10.1002/jmri.23639.
4. Mark Murphy, Marcus Alley, James Demmel, Kurt Keutzer, Shreyas Vasanawala, and Michael Lustig. Fast l1-spirit compressed sensing parallel imaging mri: Scalable parallel implementation and clinically feasible runtime. IEEE Transactions on Medical Imaging, 31(6):1250–1262, 2012. doi: 10.1109/TMI. 2012.2188039.
5. Anuroop Sriram, Jure Zbontar, Tullie Murrell, Aaron Defazio, C. Lawrence Zitnick, Nafissa Yakubova, Florian Knoll, and Patricia Johnson. End-to-end variational networks for accelerated mri reconstruction, 2020.
6. Florian Knoll, Kerstin Hammernik, Erich Kobler, Thomas Pock, Michael P Recht, and Daniel K Sodickson. Assessment of the generalization of learned image reconstruction and the potential for transfer learning. Magnetic resonance in medicine, 81(1):116–128, 2019.
7. Mohammad Zalbagi Darestani and Reinhard Heckel. Accelerated mri with un-trained neural networks. IEEE Transactions on Computational Imaging, 7:724–733, 2021.
8. Ishan Misra, Abhinav Shrivastava, Abhinav Gupta, and Martial Hebert. Cross-stitch networks for multi-task learning, 2016.
9. Alex Kendall, Yarin Gal, and Roberto Cipolla. Multi-task learning using uncertainty to weigh losses for scene geometry and semantics, 2018.
10. Michael Crawshaw. Multi-task learning with deep neural networks: A survey, 2020.
11. Simon V andenhende, Stamatios Georgoulis, Wouter Van Gansbeke, Marc Proesmans, Dengxin Dai, and Luc Van Gool. Multi-task learning for dense prediction tasks: A survey. IEEE Transactions on Pattern Analysis and Machine Intelligence, page 1–1, 2021. ISSN 1939-3539. doi: 10.1109/tpami.2021.3054719. URL http://dx.doi.org/10.1109/TPAMI.2021.3054719.
12. Xi Lin, Hui-Ling Zhen, Zhenhua Li, Qing-Fu Zhang, and Sam Kwong. Pareto multi-task learning. In H. Wallach, H. Larochelle, A. Beygelzimer, F. d'Alché-Buc, E. Fox, and R. Garnett, editors, Advances in Neural Information Processing Systems, volume 32. Curran Associates, Inc., 2019. URL https://proceedings.neurips.cc/paper/2019/file/ 685bfde03eb646c27ed565881917c71cPaper.pdf.
13. Sebastian Ruder. An overview of multi-task learning in deep neural networks, 2017.
14. Shikun Liu, Edward Johns, and A. Davison. End-to-end multi-task learning with attention. 2019 IEEE/CVFConference on Computer Vision and Pattern Recognition (CVPR), pages 1871–1880, 2019.
15. Chen Chen, Wenjia Bai, and Daniel Rueckert. Multi-task learning for left atrial segmentation on ge-mri.Lecture Notes in Computer Science, page 292–301, 2019. ISSN 1611-3349. doi: 10.1007/978-3-030-12029-0_ 32. URL http://dx.doi.org/10.1007/978-3-030-12029-0_32.
16. Kerstin Hammernik, Teresa Klatzer, Erich Kobler, Michael P. Recht, Daniel K. Sodickson, Thomas Pock, and Florian Knoll. Learning a variational network for reconstruction of accelerated mri data. Magnetic Resonance in Medicine, 79(6):3055–3071, 2018. doi: https://doi.org/10.1002/mrm.26977. URL https://onlinelibrary.wiley.com/doi/abs/10.1002/mrm.26977.
17. F Ong, S Amin, SS Vasanawala, and M Lustig. An open archive for sharing mri raw data. In ISMRM & ESMRMB Joint Annu. Meeting, page 3425, 2018.
18. Martin Uecker, Peng Lai, Mark Murphy, Patrick Virtue, Michael Elad, John Pauly, Shreyas Vasanawala, and Michael Lustig. Espirit—an eigenvalue approach to autocalibrating parallel mri: where sense meets grappa. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine, 71, 03 2014. doi: 10.1002/mrm.24751.
Figures