3925
MRI protocol recommendation using deep metric learning1Siemens Healthineers, Malvern, PA, United States
Synopsis
MRI requires careful design of imaging protocols and parameters to optimally assess a particular region of the body and/or pathological process. Selection of acquisition parameters is a challenging task because (a) the relationship between the acquisition parameters and the image features is typically non-trivial, and (b) not all users have the leverage to optimize their imaging protocols. To help users overcome these challenges and elevate the user experience, a deep metric learning tool was developed as a recommendation system for automatic candidate generation of imaging protocols. The feasibility of the model is evaluated using 3-dimensional brain MR images.
Introduction
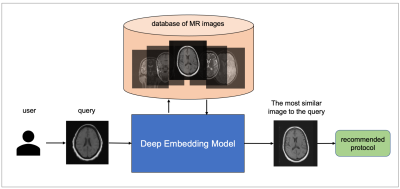
Magnetic resonance imaging requires careful design of imaging protocols and acquisition parameters to optimally assess a particular region of the body and/or pathological process. Selection of acquisition parameters is a challenging task because (a) the relationship between the acquisition parameters and the image features is typically non-trivial, and (b) not all users have the leverage to optimize their imaging protocols. Recommendation systems1 can potentially help users overcome these challenges and elevate the user experience by automatic candidate generation of the acquisition protocols. Given a query image, the system can recommend similar images to the query, together with their acquisition parameters. Deep learning-based reverse image search is one way to automate the generation of acquisition protocol from an image query selected by the user. The aim of this study is to develop a deep metric learning2 model as a recommendation system (Figure 1) for automatic candidate generation of imaging protocols, and to evaluate its feasibility on 3-dimensional brain MR images.Methods
A deep metric learning model, shown in Figure 2A, is developed for finding similar images within a database given a query. A deep convolutional neural network (CNN) maps a batch of images into an embedding space where images generated with the same acquisition parameters have smaller Euclidian distance compared to those images acquired with different parameters. A modified Google inception network3 with 3-dimensional convolution kernels is used as the deep CNN; the diagram of the network is illustrated in Figure 2B. The training was formulated as an optimization problem using the triplet loss in Equation 1:$$arg \ min_f \ max(0, \sum_{i=1}^{N}{‖f(x_i^a )-f(x_i^p )‖_2^2-‖f(x_i^a )-f(x_i^n )‖_2^2+α})$$
where is the margin, f(.) denotes embeddings, and xia, xip, xin denote the anchor, the positive, and the negative pairs respectively. An online semi-hard negative selection method4 was used to select triplets at each epoch.
The feasibility of the proposed method was evaluated on a training database constructed with 3-dimensional brain MR images. A total of 808 3-diementional T1-weighted, T2-weighted, fluid-attenuated inversion recovery (FLAIR), and time of flight (TOF), 202 images of each method, were used to construct the database. Images were sorted into different classes (protocols), where each class consists of images acquired with the exact same following acquisition parameters: repetition time (TR), echo time (TE), inversion time (IR, if applicable), scanner field strength (B0), and scanning sequence (SS). This provided 571 different protocols. To test the model, additional 400 images (100 from each imaging method) were used and were sorted into 101 different protocols.
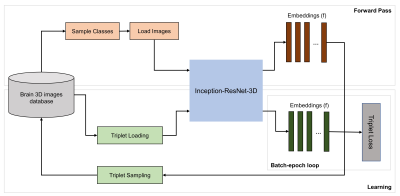
Figure 3 shows the diagram of the triplet training which consists of two phases: (1) In the forward pass, a subset of protocols is selected from the database and their corresponding embeddings are calculated. (b) In the learning phase, triplets with semi-hard negatives3 within the sampled protocols are selected and used to train the network using the triplet loss. Trainings were implemented using TensorFlow library and performed on an NVIDIA Tesla V100 GPU with the following hyperparameters: margin α=0.2, embedding size=128, size = 6, batch-epoch = 200, ADAGRAD optimizer, and 150 epochs.
The trained model was evaluated on a subset of training and test protocols. To evaluate the models on the training set, n=191 images from c=61 protocols were selected to generate np=1,525 positive and nn=1,953 negative image pairs. To evaluate on the test set, n=180 images from c=30 protocols are selected to generate np=1,427 positive and nn=1,728 negative pairs. The embedding corresponding to each image is calculated and a threshold (δ=0.4) is used to decide where the image pairs are from the same class or not. The accuracy of the model was calculated using Equation 2: $$$ accuracy=\frac{true\ positive+true\ negative}{total\ number\ of\ pairs} $$$.
Results
The accuracy of the trained model was 0.93 and 0.83 on the image pairs from training and testing sets respectively. Figure 4 illustrates the performance of the model on four example FLAIR images drawn from two protocols within the test set, the Euclidian distance between each pair, and their corresponding prediction using the deep embedding model. The model demonstrated a promising performance on mapping similar input data with similar acquisition parameters into Euclidian space where they have smaller distance compared to those acquired with different acquisition parameters.Conclusion
This feasibility study demonstrates that deep embedding learning is a promising solution for automatic protocol recommendation system. Further studies will evaluate the model on larger datasets and MR images of different anatomies.Acknowledgements
No acknowledgement found.References
1. F Araujo, R Silva, F Medeiros et al. Reverse image search for scientific data within and beyond the visible spectrum. Expert Systems with Applications. 2018.
2. M Kaya, HŞ Bilge. Deep metric learning: A survey. Symmetry. 2019
3. C Szegedy, S Ioffe, V Vanhoucke et al. Inception-v4, inception-resnet and the impact of residual connections on learning. Thirty-first AAAI conference on artificial intelligence. 2017.
4. S Florian, D Kalenichenko, J Philbin. Facenet: A unified embedding for face recognition and clustering. Proceedings of the IEEE conference on computer vision and pattern recognition. 2015.
Figures