3923
Improved Padding in CNNs for Quantitative Susceptibility Mapping1Yale University, NEW HAVEN, CT, United States
Synopsis
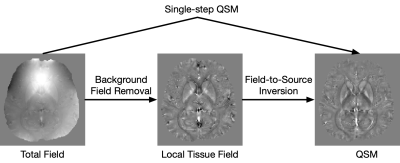
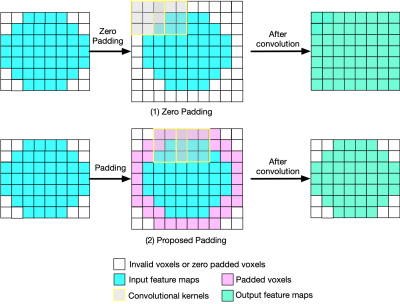
Recently, deep learning approaches have been proposed for QSM processing - background field removal, field-to-source inversion, and single-step QSM reconstruction. In these tasks, the networks usually take local fields or total fields as inputs, which have valid voxels within volume of interests (VOIs) and invalid voxels outside of VOIs. CNNs fail to consider this spatial information when using spatial invariant filters and conventional padding mechanism, which could introduce spatial artifacts in the QSM results. Here, we propose an improved padding technique utilizing neighboring valid voxels of invalid voxels to estimate the invalid voxels in feature maps of CNNs.
Introduction
Recent efforts have demonstrated the advantages of applying deep learning (DL) for quantitative susceptibility mapping (QSM) in background field removal[1,2], field-to-source inversion[3,4,5], and single-step QSM[6]. All these methods utilized U-Net like convolutional neural networks (CNNs). In these tasks, the networks usually take local fields or total fields as inputs, which have valid voxels within volume of interest (VOI) and invalid voxels outside of VOI. CNNs fail to consider this spatial information when using spatial invariant filters and conventional padding mechanism, which could introduce spatial artifacts in the network inference. Here, we propose an improved padding technique utilizing neighboring valid voxels of invalid voxels to estimate the invalid voxels in feature maps of CNNs.Method
Let $X$ are the feature values (voxels values) and $M$ is the corresponding binary mask. First, a convolution with all-one 3x3x3 kernel was the padded binary mask to get the scaling factor $1/sum(M)$ which applies appropriate scaling to adjust for the varying amount of invalid inputs. Second, a convolution with all-one 3x3x3 kernel was the each feature map to get the average value of valid neighboring voxels for the invalid voxels. For better generalization, the convolution kernels for feature maps and binary mask were set as trainable parameters, which were initialized with value of one.We used the COSMOS result of 2016 QSM reconstruction challenge[7] to generate the simulated data. We applied random elastic transform, contrast change, and adding pseudo high susceptibility sources to augment the single QSM. The background field were simulated by placing random background susceptibility sources with large susceptibility values outside the brain. The dipole convolution were then performed to get the induced total field and local field from the susceptibility distribution. 100 datasets with matrix size 160x160x160 and voxel size 1.0x1.0x1.0mm$^3$ were generated for network training tasks for background field removal, field-to-source inversion, and single-step QSM. The network adopted a 3D U-Net like architecture, using patch-based training with patch size 96x96x96, and L2 loss. We compared four padding techniques - (1) zero padding, (2) reflective padding, (3) symmetric padding, and (4) the proposed one. 100 testing datasets were generated using the same way as training data. The prediction results were evaluated with respect to the ground truth using quantitative metrics - peak signal to noise ratio (PSNR), normalized root mean squared error (NRMSE), high frequency error norm (HFEN), and structure similarity index (SSIM).
9 QSM datasets were acquired using 5 head orientations and a 3D single-echo GRE scan with isotropic voxel size 1.0x1.0x1.0 mm$^3$ on 3T MRI scanners. QSM data processing was implemented as following, offline GRAPPA reconstruction to get magnitude and phase images from saved k-space data, coil combination using sensitivities estimated with ESPIRiT BET (FSL, Oxford, UK) for brain extraction, Laplacian method for phase unwrapping, and RESHARP[8] with spherical mean radius 4mm for background field removal. COSMOS results were calculated using the 5 head orientation data which were registered by FLIRT (FSL, Oxford, UK). In network training for background field removal, the RESHARP results were used as the training label. Since RESHARP local fields have brain erosion, the input total field used the eroded volume. Leave-one-out cross validation was used. For each dataset, total 40 scans (8*5) from other 8 datasets were used for training. The network was trained on patch-based with patch size 96x96x96, L2 loss. For field-to-source inversion task, the COSMOS maps were used as training label with network input of local tissue field. Leave-one-out cross validation was used, only using the normal head position scans for training and testing. For single-step QSM reconstruction task, the COSMOS maps were used as the training label with the total field as the network input. Leave-one-out cross validation was used, only using the normal head position scans for training and testing.
Result
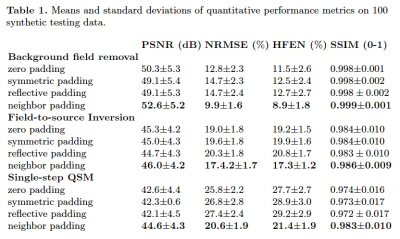
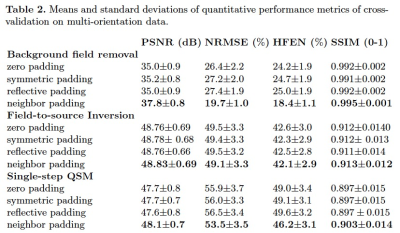
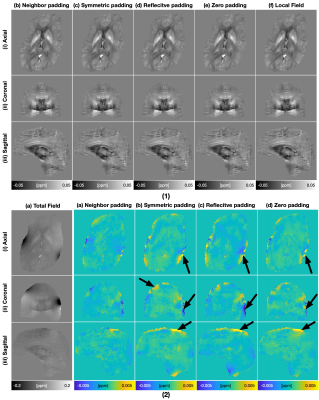
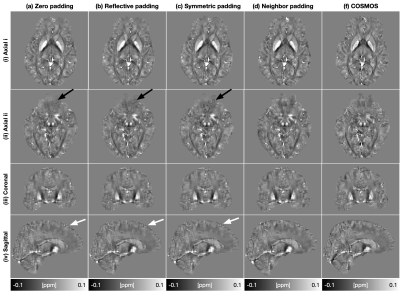
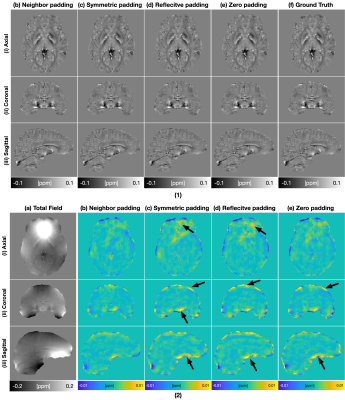
Table.1 displays the quantitative evaluation results. In the three tasks, the proposed method achieved the best scores in all metrics. Fig.3 displays the background field removal results of a synthetic test data using different padding techniques. From the residual maps, the proposed padding technique have obvious less residual errors, especially close to brain boundaries where have a strong background field.Table.2 lists the quantitative evaluation results. In all three tasks, the proposed method achieved the best scores across all metrics. Fig.4 shows that the single-step QSM results of conventional padding techniques could introduce artifacts at the brain boundaries especially at regions with strong background fields, while the proposed method could suppress these artifacts. Fig.5 displays the background field removal results from networks with different padding techniques. From the residual error maps, the result of the proposed padding technique have obvious residual errors especially close to brain boundaries.
Discussion and Conclusion
In this work, an improved padding technique was proposed to decrease the spatial artifacts in QSM CNNs. From quantitative evaluation and visual assessment on synthetic datasets and in-vivo datasets, the proposed padding technique achieved impressive performance with substantial less errors than conventional padding techniques, especially in the tasks of background field and single-step QSM. We believe that the proposed padding technique could improve DL-based QSM techniques.Acknowledgements
We thank Professor Jongho Lee for sharing the multi-orientation QSM datasets.References
1. Bollmann, Steffen, Matilde Holm Kristensen, Morten Skaarup Larsen, Mathias Vassard Olsen, Mads Jozwiak Pedersen, Lasse Riis Østergaard, Kieran O’Brien, Christian Langkammer, Amir Fazlollahi, and Markus Barth. "SHARQnet–Sophisticated harmonic artifact reduction in quantitative susceptibility mapping using a deep convolutional neural network." Zeitschrift für Medizinische Physik 29, no. 2 (2019): 139-149.
2. Liu, Juan, and Kevin M. Koch. "Deep Gated Convolutional Neural Network for QSM Background Field Removal." In International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 83-91. Springer, Cham, 2019.
3. Yoon, Jaeyeon, Enhao Gong, Itthi Chatnuntawech, Berkin Bilgic, Jingu Lee, Woojin Jung, Jingyu Ko et al. "Quantitative susceptibility mapping using deep neural network: QSMnet." Neuroimage 179 (2018): 199-206.
4. Bollmann, Steffen, Kasper Gade Bøtker Rasmussen, Mads Kristensen, Rasmus Guldhammer Blendal, Lasse Riis Østergaard, Maciej Plocharski, Kieran O'Brien, Christian Langkammer, Andrew Janke, and Markus Barth. "DeepQSM-using deep learning to solve the dipole inversion for quantitative susceptibility mapping." Neuroimage 195 (2019): 373-383.
4. Jung, Woojin, Jaeyeon Yoon, Sooyeon Ji, Joon Yul Choi, Jae Myung Kim, Yoonho Nam, Eung Yeop Kim, and Jongho Lee. "Exploring linearity of deep neural network trained QSM: QSMnet+." Neuroimage 211 (2020): 116619.
5. Chen, Yicheng, Angela Jakary, Sivakami Avadiappan, Christopher P. Hess, and Janine M. Lupo. "QSMGAN: improved quantitative susceptibility mapping using 3D generative adversarial networks with increased receptive field." NeuroImage 207 (2020): 116389.
6. Wei, Hongjiang, Steven Cao, Yuyao Zhang, Xiaojun Guan, Fuhua Yan, Kristen W. Yeom, and Chunlei Liu. "Learning-based single-step quantitative susceptibility mapping reconstruction without brain extraction." NeuroImage 202 (2019): 116064.
7. Langkammer, Christian, Ferdinand Schweser, Karin Shmueli, Christian Kames, Xu Li, Li Guo, Carlos Milovic et al. "Quantitative susceptibility mapping: Report from the 2016 reconstruction challenge." Magnetic resonance in medicine 79, no. 3 (2018): 1661-1673.
8. Sun, Hongfu, and Alan H. Wilman. "Background field removal using spherical mean value filtering and Tikhonov regularization." Magnetic resonance in medicine 71, no. 3 (2014): 1151-1157.
Figures