3921
Exploration of vision transformer models in medical images synthesis1Department of Electrical and Computer Engineering, University of Wisconsin-Madison, Madison, WI, United States, 2Department of Radiology, University of Wisconsin-Madison, Madison, WI, United States, 3Department of Biomedical Engineering, Department of Radiology and Medical Physics, Madison, WI, United States
Synopsis
Applications such as PET/MR and MR-only Radiotherapy Planning need the capability to derive a CT-like image from MRI inputs to enable accurate attenuation correction and dose estimation. More recently, transformer models have been proposed for computer vision applications. Models in the transformer family discard traditional convolution-based network structures and emphasize the importance of non-local information yielding potentially more realistic outputs. To evaluate the performance of SwinIR, TransUet, and Unet. After comparing results visually and quantitatively, the SwinIR models and TransUnet models appear to provide higher-quality synthetic CT scans compared to the conventional Unet.
Introduction
Applications such as PET/MR and MR-only Radiotherapy Planning need the capability to derive a CT-like image from MRI inputs to enable accurate attenuation correction and dose estimation. Recently, many successful implementations using deep learning have been presented [1]. Due to the rapid growth of deep learning-based approaches, many distinct architectures have been explored. More recently, transformer models have been proposed for computer vision applications. The transformer model is originally designed for natural language processing and its architecture has been adapted into the vision transformer model (ViT) [2]. The ViT model splits the input images into a sequence of patches, and a self-attention mechanism is employed to learn the intrinsic relationship between patches. Models in the ViT family discard traditional convolution-based network structures and emphasize the importance of non-local information yielding potentially more realistic outputs. Recall the highly popular Unet model[3], containing an encoder-decoder structure that implements convolution-based blocks to encode and decode data features. Recently, the TransUnet [4] model has been proposed with a similar Unet structure, but uses ViT modules as the encoders, tokenizing image patches as the input for extracting hidden features with convolutional decoding blocks. The Swin [5] ViT model shifts the patch partitioning window from fixed grids into sliding grids, increasing efficiency by allowing for cross-patch connections. Furthermore, SwinIR [6] adds several convolutional layers to extract the shallow features leading to the reconstructions of high-quality images. The purpose of this research is to compare these highly novel transformer architectures to a more conventional Unet in a MR to CT translation task.Methods
IRB-approved retrospective clinical image data were obtained from 64 patients at our institution who had undergone a clinical CT and MRI scan on the same day. MR imaging was performed on GE Healthcare 1.5T MRI scanners using a T1-weighted gradient echo (T1) acquisition (BRAVO, TE = ∼3.5 ms, TI = 450 ms, TR = ∼10 ms, 1 × 1 × 2.4–3.0 mm resolution). Images were registered pairwise using AntsPy (http://www.picsl.upenn.edu/ANTS/) and both images were approximately registered to the MNI space using an affine transformation computed from the T1 image and resampled to 1 x 1 x 1 mm resolution. To evaluate the performance of SwinIR, TransUet, and Unet, data was divided as follows; 31 subjects used for training, 8 subjects considered for validation, and 25 subjects used for testing. Due limitted of GPU memory, the training pairs were down-sampled to one-half the resolution for the SwinIR image translation model. Following training, a SwinIR-based super-resolution network was cascaded to upsampled the output into the original space. Image translation using the SwinIR model was refined on the study data from a pre-trained model originally used for denoising color images while the super-resolution SwinIR was pre-trained using the DIV2K dataset. The transformer portion of the TransUnet model was pre-trained on the ImageNet dataset. It should be mentioned that the decoder part of TransUnet and the Unet model were trained from the scratch. All networks utilized the smooth L1-norm loss function and AdamW optimizer.Results
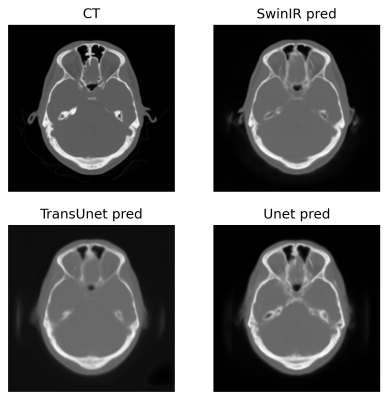
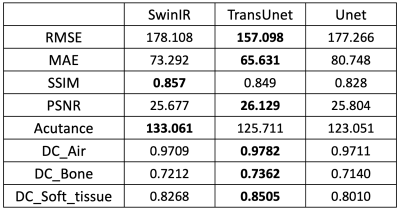
To compare the image quality of synthetic CTs produced by SwinIR, TransUet and Uet, the root of mean squared error (RMSE), mean absolute error (MAE), structural similarity index (SSIM), and peak signal-to-noise ratio (PSNR) were calculated, and the results are shown in Table 1. A paired t-test between SwinIR vs. Unet and TransUnet vs. Unet yielded no statistically significant differences. Acutance was also calculated as the mean of the absolute sum of gradients. Acutance is the metric of sharpness and the higher the acutance is, the sharper the image is. Additionally, the dice coefficient (DC) were used as the measure of overlap between regions of a segmented CT image. To do so, air, bone and soft tissue were thresholded according to the HU value (<-500, >500 and [-200, 200]), for evaluating the DC.Overall, the TransUnet performs best for all metrics except SSIM and Acutance. In Figure 1, the SwinIR model gives the sharpest images as suggested by its higher Acutance.
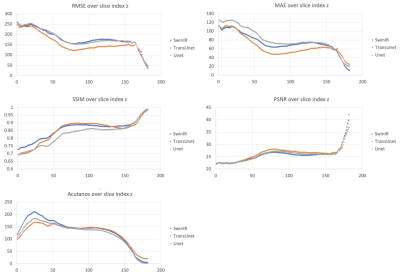
The distribution of metrics (RMSE, MAE, SSIM, PSNR, Acutance) across axial slices in the test dataset is depicted in Figure 2 where lower slice numbers are the more inferior portion of the head.
Discussion and Conclusion
After comparing results visually and quantitatively, the SwinIR models and TransUnet models appear to provide higher-quality synthetic CT scans compared to the conventional Unet. Future developments in specifically-designed vision transformer models, specifically 3D methods, are expected to achieve better performance than demonstrated herein.Acknowledgements
No acknowledgement found.References
1. Spadea M F, Maspero M, Zaffino P, et. Deep learning-based synthetic-CT generation in radiotherapy and PET: a review. arXiv preprint arXiv:2102.02734; 2021.
2. Dosovitskiy A, Beyer L, Kolesnikov A, et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:2010.11929; 2020.
3. Ronneberger O, Philipp F, Thomas B. U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical image computing and computer-assisted intervention. Springer, Cham; 2015.
4. Chen J, Lu Y, Yu Q, et al. Transunet: Transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:2102.04306; 2021.
5. Liu Z, Lin Y, Cao Y, et al. Swin transformer: Hierarchical vision transformer using shifted windows. arXiv preprint arXiv:2103.14030; 2021.
6. Liang J, Cao J, Sun G, et al. SwinIR: Image restoration using swin transformer. Proceedings of the IEEE/CVF International Conference on Computer Vision (pp. 1833-1844); 2021.
Figures