3847
Single-offset and multi-offset super-resolution for CEST MRI using deep transfer learning1Hong Kong Centre For Cerebro-Cardiovascular Health Engineering, Hong Kong, Hong Kong, 2Department of Biomedical Engineering, City University of Hong Kong, Hong Kong, Hong Kong, 3Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 4City University of Hong Kong Shenzhen Research Institute, Shenzhen, China
Synopsis
CEST MRI is an unique molecular imaging approach to reveal the exchangeable proton information related to physiology and pathology. However, long scanning time has hindered its translation into clinics. While deep-learning based super-resolution methods have been explored to reduce scanning time in conventional MRI, adaptation of these methods to CEST MRI has been limited due to lack of large public CEST datasets. Therefore, this study proposes two transfer learning based super-resolution methods, Single-Offset UNet and Multi-Offset UNet, for accelerating CEST MRI acquisition by using public MRI databases for pretraining and a very small CEST dataset for training.
INTRODUCTION
Rapid development in precision medicine has led to increasing demand for clinical imaging modalities that provide molecular and physiological information, in addition to anatomical features1,2. Chemical Exchange Saturation Transfer (CEST) MRI, with its molecular imaging capabilities, shows promising applications in early disease diagnosis and guided treatment3,4. However, long scanning time for CEST MRI is a constraint in its clinical applications5, and therefore, requires a compromise between scanning time and image quality6. Deep-Learning-based Super-Resolution (DL-SR) methods have been explored in conventional MRI for reconstructing high-resolution images from low-resolution acquisitions, which leads to a shorter acquisition time6. The lack of large public CEST databases for DL-SR model development poses an obstacle in adapting these methods to CEST MRI. Hence, this study aims to tackle this issue by proposing two DL-SR methods, Single-Offset UNet (SO-UNet) and Multi-Offset UNet (MO-UNet), for accelerating CEST MRI acquisition using large public MRI datasets for pretraining, facilitating the training of the neural networks with very small CEST datasets.METHODS
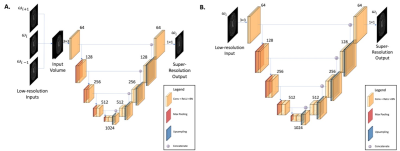
Data Preparation:For pretraining, human brain MRI images from TCIA database LGG-1p19qDeletion study7-9 were selected and resized to 96×96 matrices using Lanczos downsampling to prevent scale variance10 between pretraining and training dataset. For each high-resolution image, the corresponding low-resolution MRI image was generated by downsampling of the K-space, followed by inverse Fourier transformation11 (Figure 1).
For training, 43 CEST MRI datasets of mouse brain (C57/BL6) were used. The corresponding low-resolution CEST MRI images were generated by the downsampling method indicated in Figure 1.
Model:
Two modified iterations of U-Net12, referred to as SO-UNet and MO-UNet, were developed for reconstructing high-resolution CEST images (96×96) from low-resolution CEST images (48×48) (Figure 2).
Model Development:
Both pretraining and training data were randomly split into 80% for training (13888 for pretrain and 3163 for train datasets) and 20% for validation (3472 for pretrain and 791 for train datasets). Due to the small training dataset size, the mouse CEST images were augmented to increase variability in the dataset.
Both models were trained on the pretraining data for 1000 epochs to initialize the weights of the models, following which, were trained on training data for 1500 epochs to fine-tune the weights.
Both SO-UNet and MO-UNet were optimized on a hybrid loss function composed of L1 loss, multi-scale similarity index measure (MS-SSIM) loss13 and edge loss14
$$Loss^{hybrid} = \alpha \cdot Loss^{ms-ssim} + \beta \cdot Loss^{edge} + (1- \alpha - \beta) \cdot Loss^{L1}$$
where $$$ \alpha$$$ = 0.7, $$$\beta$$$ = 0.2.
Performance Evaluation:
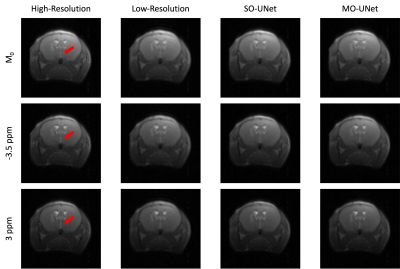
To evaluate the performance of the model, CEST MRI acquisition of 2 brain tumour mice (NOD-SCID), that were not part of the training process, were used. The corresponding low-resolution CEST images were generated using the same method (Figure 1).
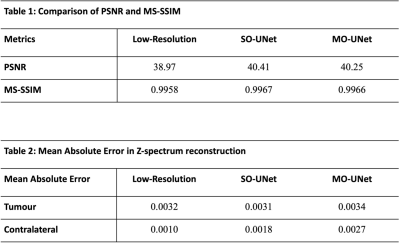
Both MO-UNet and SO-UNet were evaluated based on two criteria: performances in reconstructing spatial features and reconstructing Z-spectrum. To quantify the performance in reconstructing spatial features, Peak Signal-to-Noise ratio (PSNR)15 and MS-SSIM15,16 were calculated. PSNR measures the pixel-level similarity while MS-SSIM measures the structural similarity between two images.
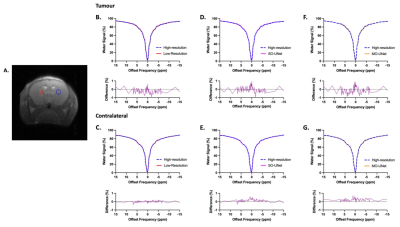
To evaluate the performance in reconstructing the Z-spectrum, the Mean Absolute Error (MAE) of the Z-spectra from the tumour and its contralateral side of the high-resolution CEST dataset were compared with the same region of interests (ROIs) in low-resolution, SO-UNet, and MO-UNet CEST datasets.
RESULTS AND DISCUSSION
From Figure 3, it is evident that both SO-UNet and MO-UNet performed well in reconstructing high-frequency features, such as the region indicated by arrow, that were not captured in the low-resolution images. Results in Table 1 show that both models improved the spatial resolution, reflected by the higher PSNR and MS-SSIM of both models’ outputs compared with low-resolution images. Between the two models, the SO-UNet marginally outperformed the MO-UNet on both metrics. A possible reason could be that both models were not trained to their optimal performance and hence further investigation is required.When reconstructing the Z-spectra (Table 2), it is evident that both models performed well, indicated by the low MAE which is insignificant to cause deviation in the Z-spectrum. Therefore, the reconstructed Z-spectra retains the molecular information present in high-resolution Z-spectra. Notably, it can be observed that the Z-spectra from low-resolution datasets also have a low MAE. This can be attributed to the fact that the central window (48×48) used in the downsampling method to generate the low-resolution images preserved most of the pixel information that is used to construct the Z-spectrum. However, a smaller central window, such as 32×32, used in the downsampling method might lead to larger pixel information loss, causing deviation in Z-spectrum, which needs to be further investigated.
CONCLUSION
In this study, we proposed the use of two DL-SR methods for reconstructing high-resolution CEST images from low-resolution acquisitions, and thereby, shortening the scanning time. We also illustrated that the DL-SR models can achieve promising results on a small CEST MRI dataset by taking advantage of pretraining on public MRI databases. Further investigations are underway to evaluate its ability to reconstruct high-resolution images from smaller image acquisition, such as 32×32. Current results indicate that deep-learning-based SO-UNet and MO-UNet show promise in shortening CEST MRI scanning time without compromise in spatial and spectral quality, which facilitates the clinical translation of CEST MRI.Acknowledgements
This work was supported by Research Grants Council: 11102218, PDFS2122-1S01; City University of Hong Kong: 7005210, 7005433, 9680247, 9667198 and 9609307; National Natural Science Foundation of China: 81871409.References
1. Ghasemi M, Nabipour I, Omrani A, Alipour Z, Assadi M. Precision medicine and molecular imaging: new targeted approaches toward cancer therapeutic and diagnosis. Am J Nucl Med Mol Imaging 2016;6(6):310-327.
2. Giardino A, Gupta S, Olson E, Sepulveda K, Lenchik L, Ivanidze J, Rakow-Penner R, Patel MJ, Subramaniam RM, Ganeshan D. Role of Imaging in the Era of Precision Medicine. Acad Radiol 2017;24(5):639-649.
3. Jones KM, Pollard AC, Pagel MD. Clinical applications of chemical exchange saturation transfer (CEST) MRI. J Magn Reson Imaging 2018;47(1):11-27.
4. van Zijl PC, Yadav NN. Chemical exchange saturation transfer (CEST): what is in a name and what isn't? Magn Reson Med 2011;65(4):927-948.
5. Villano D, Romdhane F, Irrera P, Consolino L, Anemone A, Zaiss M, Dastru W, Longo DL. A fast multislice sequence for 3D MRI-CEST pH imaging. Magn Reson Med 2021;85(3):1335-1349.
6. Plenge E, Poot DH, Bernsen M, Kotek G, Houston G, Wielopolski P, van der Weerd L, Niessen WJ, Meijering E. Super-resolution methods in MRI: can they improve the trade-off between resolution, signal-to-noise ratio, and acquisition time? Magn Reson Med 2012;68(6):1983-1993.
7. Erickson B, Akkus Z, Sedlar J, Kofiatis P. Data from LGG-1p19qDeletion. The Cancer Imaging Archive 2017;76.
8. Akkus Z, Ali I, Sedlar J, Agrawal JP, Parney IF, Giannini C, Erickson BJ. Predicting Deletion of Chromosomal Arms 1p/19q in Low-Grade Gliomas from MR Images Using Machine Intelligence. J Digit Imaging 2017;30(4):469-476.
9. Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. Journal of digital imaging 2013;26(6):1045-1057.
10. Goodfellow I, Bengio Y, Courville A. Deep learning: MIT press; 2016.
11. Masutani EM, Bahrami N, Hsiao A. Deep Learning Single-Frame and Multiframe Super-Resolution for Cardiac MRI. Radiology 2020;295(3):552-561.
12. Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. 2015. Springer. p 234-241.
13. Zhao H, Gallo O, Frosio I, Kautz J. Loss functions for neural networks for image processing. arXiv preprint arXiv:151108861 2015.
14. Seif G, Androutsos D. Edge-based loss function for single image super-resolution. 2018. IEEE. p 1468-1472.
15. Wang Z, Chen J, Hoi SCH. Deep Learning for Image Super-Resolution: A Survey. IEEE Trans Pattern Anal Mach Intell 2021;43(10):3365-3387.
16. Wang Z, Simoncelli EP, Bovik AC. Multiscale structural similarity for image quality assessment. 2003. Ieee. p 1398-1402.
Figures