3837
Comparison of 3D amide proton transfer (APT) and four advanced diffusion models in gliomas IDH status decoding1Department of Radiology,The Second Xiangya Hospital, Central South University, Changsha, China, 2MR Scientific Marketing, Siemens Healthineers, Wuhan, China, 3MR Application, Siemens Healthineers, Changsha, China, 4Shanghai Key Laboratory of Magnetic Resonance, School of Physics and Electronic Science, Shanghai Key Laboratory of Magnetic Resonance, School of Physics and Electronic Science,East China Normal University, Shanghai, China
Synopsis
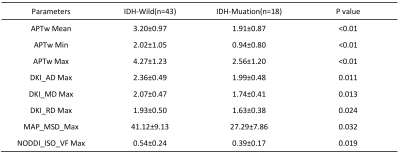
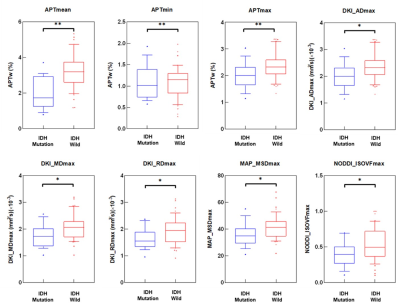
The purposed of study was to compare the diagnostic efficacy of APT imaging and four diffusion models, including DTI, DKI, NODDI, and MAP, in evaluating genotype IDH status, and to find the best imaging indicators for aiding accurate diagnoses and treatment decisions. Compared with IDH-mutant gliomas, IDH-wildtype gliomas had significantly higher mean and maximum of APTw, maximum axial, mean and radial diffusivity from DKI, maximum mean square diffusivity from MAP and maximum isotropic volume fraction from NODDI, as well as lower minimum APTw value.
Introduction
Gliomas account for approximately 77% of primary malignant brain tumors.1 The classification of gliomas has been mainly based on concepts of histogenesis. Recent advancements in neuro-oncology have changed the focus away from histopathologic grading and toward molecular characteristics that have been incorporated into the WHO classification. Especially for diffuse astrocytic and oligodendroglial tumors, isocitrate dehydrogenase (IDH) genotypes are important for subtyping.2 The most crucial information asked by neuroradiologists is not only grading but also molecular characteristics, especially in nonenhancing lower-grade gliomas. Imaging must adapt to this paradigm shift to remain relevant, and its job should be repurposed to identify molecular status. Amide proton transfer weighted (APTw) MRI, which is a new MR technique with demonstrated potential ability of tumor grading and IDH mutation status prediction by reflecting biologically active tumor portion that has high cellularity and proliferation at histopathologic correlation.3-5 Diffusion-weighted imaging (DWI) is a valuable imaging biomarker for classifying gliomas because it allows tumor characteristics to be measurable without the need of tracer injections. Recently, novel MRI diffusion models, such as diffusion kurtosis imaging (DKI), neurite orientation dispersion and density imaging (NODDI), and the non-Gaussian-based mean apparent propagator (MAP)-MRI, which can provide insights into the brain microstructure, have become the preferred methods for analyzing gliomas tissues.6-7 Our goal was to compare the diagnostic efficacy of four diffusion models (DTI, DKI, NODDI, MAP) and APT imaging in evaluating genotype IDH status, and to find the best imaging indicators for aiding accurate diagnoses and treatment decisions.Materials and Methods
The patients with histopathologically proven gliomas between May 2020 and Aug 2021 were included, and they met the following inclusion criteria: (1) MRI scans had been performed before previously untreated; (2) patients with the IDH mutation status of genotype was acquired. Finally, a total of 61 patients (male/female, 37/25; mean age, 51.9 ± 13 years, 43 patients with IDH-Wild,18 patients with IDH-Mutant) were included. MR imaging examinations were performed on a 3T scanner (MAGNETOM Skyra; Siemens Healthcare, Erlangen, Germany) with a 32-channel head coil. The conventional sequences included axial pre-contrast T2 FLAIR-weighted TSE sequence, pre- and postcontrast T1-weighted MPRAGE sequence, and DWI and APTw images were acquired before contrast agent administration. The APT imaging based on a novel whole-brain isotropic-resolution CEST sequence, and parameters were as follows: FOV = 212×212×201 mm3, matrix = 76×76×72, resolution = 2.8×2.8×2.8mm3, TR = 3 seconds, TE = 17ms, turbo factor = 140, number of averages = 1.2, and GRAPPA factor = 2×2, scan time=4 min38s. Seven CEST saturation offsets for APT-weighted (APTw) imaging were executed, including unsaturated (S0) and saturated frequencies of ±3 ppm, ±3.5 ppm, and ±4 ppm. Diffusion MRI was performed in the transverse plane using a half q-space Cartesian grid diffusion model with a radial grid size of 3. The following parameters was used: TR/TE= 4500/111 ms; 60 slices; FOV= 192×192 mm2, resolution = 2.0× 2.0×2.0 mm3, and b=0, 350, 650, 1000, 1350, 1650, 2000, 2650, 2700, 3000 s/mm2, scan time=5min12s. The T-test or Mann-Whitney U test were used to compare IDH-Wild and IDH-Mutation. The receiver operating curve (ROC) was utilized to calculate the sensitivity, specificity, area under the curve (AUC). P < 0.05 was considered statistically significant.Results
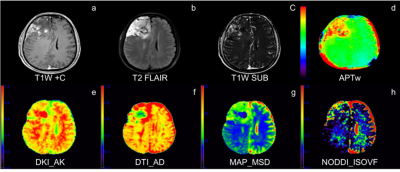
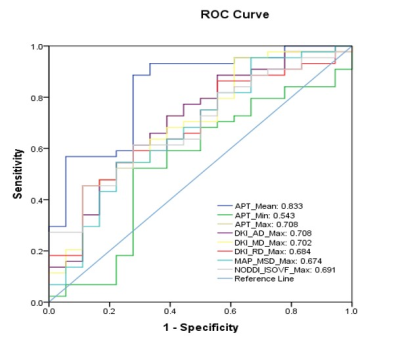
Table 1 and Figure 1 shows the parameters that have significant differences between the IDH-Wild and IDH-Mutation. Compared with IDH-mutant gliomas, IDH-Wild gliomas showed significantly higher mean APTw, maximum values of APTw, axis, radial and mean diffusivity from DKI (DKI_AD, DKI_MD, DKI_RD), mean squared displacement from MAP (MAP_MSD) and isotropic volume fraction from NODDI (NODDI_ISOVF), as well as lower minimum APTw value (p<0.05). The parameter maps of one patient with IDH-Mutation are shown in Figure 2.Figure 3 shows the ROC in differentiating IDH-Mutation and IDH-Wild for the parameters in Table 1, and the associated area under the curve, accuracy, sensitivity, specificity, and positive and negative predictive values are shown in Table 2. The 8 metrics had different areas under the curve, ranging from 0.543 to 0.833. Mean value of APTw had the largest AUC (0.833).
Discussion
Regarding the APT signals of IDH genotype prediction, we found that mean APTw is significant higher in IDH-Wild genotype (AUC 0.833; best cut-off value 2.10%; p<0.01). According to the CEST theory, APT MRI can provide contrast correlated with metabolite concentrations and tumor cellularity based on the cellular mobile proteins and peptides.8 The diagnostic performance of our present study in the larger cohort agreed with the prediction by previous studies reported.9 This study used a novel whole-brain isotropic CEST sequence using SPACE method. This new CEST-SPACE technique can rapidly generate whole-brain CEST source images with negligible susceptibility artifact, which might improve the image quality of APTw. This impacts clinical applications about APT because SPACE metrics can acquire better images in less time than EPI acquisition.Conclusion
The results revealed that APT has higher diagnostic accuracy than DTI, DKI, MAP and NODDI, implying that APTw is a reliable biomarker for glioma classification.Acknowledgements
No acknowledgement found.References
[1] HÜBNER J, KOOL M, PFISTER S, et al. Epidemiology, molecular classification and WHO grading of ependymoma [J]. Journal of neurosurgical sciences, 2018, 62(1): 46-50.
[2] LOUIS D N, PERRY A, REIFENBERGER G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary [J]. Acta Neuropathol, 2016, 131(6): 803-20.
[3] ZHOU J, LAL B, WILSON D A, et al. Amide proton transfer (APT) contrast for imaging of brain tumors [J]. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine, 2003, 50(6): 1120-6.
[4] JIANG S, ZOU T, EBERHART CG, et al. Predicting IDH mutation status in grade II gliomas using amide proton transfer-weighted (APTw) MRI [J]. Magn Reson Med, 2017, 78(3): 1100-9.
[5] ZHOU J, ZHU H, LIM M, et al. Three-dimensional amide proton transfer MR imaging of gliomas: Initial experience and comparison with gadolinium enhancement [J]. J Magn Reson Imaging, 2013, 38(5): 1119-28.
[6] JIANG R, JIANG S, SONG S, et al. Laplacian-Regularized Mean Apparent Propagator-MRI in Evaluating Corticospinal Tract Injury in Patients with Brain Glioma [J]. Korean journal of radiology, 2020, [7] CHU J, SONG Y, TIAN Y, et al. Diffusion kurtosis imaging in evaluating gliomas: different region of interest selection methods on time efficiency, measurement repeatability, and diagnostic ability [J]. European radiology, 2020.
[8] ZHOU J, PAYEN J-F, WILSON D A, et al. Using the amide proton signals of intracellular proteins and peptides to detect pH effects in MRI [J]. Nature medicine, 2003, 9(8): 1085-90. [9] TOGAO O, HIWATASHI A, YAMASHITA K, et al. Grading diffuse gliomas without intense contrast enhancement by amide proton transfer MR imaging: comparisons with diffusion-and perfusion-weighted imaging [J]. European radiology, 2017, 27(2): 578-88.
Figures