3820
BUDA-SAGE with Slider encoding and self-supervised denoising enables fast, distortion-free, high-resolution T2 and T2* mapping1State Key Laboratory of Modern Optical Instrumentation, College of Optical Science and Engineering, Zhejiang University, Hangzhou, China, 2Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Boston, MA, United States, 3Subtle Medical Inc, Menlo Park, CA, United States, 4Radiological Sciences Laboratory, Stanford University, Stanford, CA, United States, 5Department of Radiology, Harvard Medical School, Boston, MA, United States, 6Harvard-MIT Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA, United States
Synopsis
BUDA-SAGE is an efficient multi-shot echo‐planar imaging (EPI) sequence that provides multiple distortion-free T2*-, T2’- and T2-weighted contrasts for quantitative mapping. In this work, we employ Slider encoding to reach 1 mm isotropic resolution by performing super-resolution reconstruction on volumes acquired with 2 mm slice thickness. A self-supervised neural network MR-Self2Self (MR-S2S) was utilized to perform denoising to boost SNR. Estimated quantitative maps showed consistent values with conventional mapping methods in phantom and in-vivo measurements. We demonstrate that BUDA-SAGE acquisition with self-supervised denoising and Slider encoding enables rapid, distortion-free, whole-brain T2/T2* mapping at 1 mm isotropic resolution under 90 seconds.
Introduction
Quantitative parameter mapping has demonstrated potential in clinical and neuroscience applications (1), but its adoption has been hampered by the long acquisition time required to encode multi-contrast images that capture the signal evolution. Many emerging methods including MR fingerprinting(2,3), MR Multitasking (4) and 3D-EPTI (5) have been developed to improve the efficiency and reliability of quantitative imaging.Recently, a new method termed BUDA-SAGE(Blip Up-Down Acquisition-Spin And Gradient Echo) (6) was proposed, which utilizes alternate phase-encoding polarity across the interleaved shots and multiple EPI readouts with self-supervised MR-S2S denoising to achieve rapid and distortion-free multi-parametric T2/T2* mapping with high SNR. Super-resolution techniques (7,8) such as Slider (SLIce Dithered Enhanced Resolution) have shown promise for increasing image resolution while retaining high SNR. Inspired by Slider, we propose to acquire a number of multislice data sets, each volume shifted by a sub-pixel amount in the slice direction with respect to the other volumes, and perform super-resolution reconstruction to achieve high isotropic resolution quantitative mapping.
By combining BUDA-SAGE with Slider and MR-S2S, we propose to harness the efficiency of this distortion-free acquisition in high-resolution imaging with high SNR, and demonstrate whole-brain T2/T2* maps at 1mm isotropic resolution under 90 seconds. Phantom and in-vivo results show that the estimated maps are consistent with the gold standard mapping acquisitions. BUDA-SAGE reduces the scan time dramatically for increased motion robustness, while providing clinically desired, multi-contrast data and multi-parameter maps.
Methods
Acquisition: Data were collected on a 3T Siemens Prisma using 32-channel reception with the approval of Institutional Review Board.(A) To validate the accuracy of T2/T2* maps, BUDA-SAGE data were collected using the following protocol: resolution=1×1×2mm3, FOV=220×220×120mm3, Rin-plane=8, SMS factor=2 with 8-shots. We changed the TEs and performed three separate scans to acquire three groups of data with [TEgre, TEmixed, TEse]=[18, 64, 91] in group1, [30, 88, 115] in group2 and [42, 112, 139] in group3 at TR=5s. Total acquisition time=125s(Tacq=TR×Nshot×Ngroup+Tdummy=5×8×3+5).
For conventional mapping acquisitions, to quantify T2* map, a multi-echo GRE sequence with 10 different TEs(from 6ms to 60ms with a gap of 6ms), TR=5s, resolution=1×1×2mm3, FOV=220×220×120mm3 was acquired in a scan time of 8min 35s. To derive a T2 map, six multi-slice single spin echo acquisitions with different TEs(from 19ms to 139ms with a gap of 24ms), TR=7s, resolution=1×1×2mm3, FOV=192×174×120mm3 were acquired in a scan time of 40min 36s.
(B) To perform Slider super-resolution and provide 1mm isotropic resolution of T2/T2* maps, two groups (group1 and group3) and 4-shots of BUDA-SAGE data were acquired on a second volunteer. The acquisition parameters were the same as (A). We collected each of these two groups data at the slice thickness of 2mm. We performed an additional acquisition by shifting the slice position by 1mm for each group. The total acquisition time=85s(Tacq=TR×Nshot×Ngroup×Nslider+Tdummy=5×4×2×2+5).
An FOV-matched low resolution 2D GRE was acquired to obtain distortion-free sensitivity maps for all BUDA-SAGE reconstructions from a 2s calibration scan.
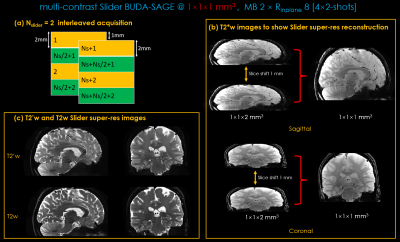
Slider super-resolution reconstruction: Slider acquires Nslider sets of high SNR, thick-slice data (at resolution 1×1×Nslider mm3) with 1mm shift between each set along the slice direction to achieve 1mm isotropic resolution final results. The Nslider=2 interleaved acquisition is displayed on Figure 4(a). To achieve thin-slice high-resolution reconstruction, data were first reconstructed using BUDA-SAGE reconstruction with MR-S2S denoising. The thin slices are then resolved within the acquired thick-slice images using linear reconstruction with Tikhonov regularization:
$$z_{rec} = (A^T A + \lambda_{Tik} I)^{-1} A^T b = A_{inv_{\_Tik}}\;b$$
Where A is the Slider encoding matrix that sums adjacent slices, b is the concatenation of the acquired Slider-encoded thick slice images, zrec is the corresponding high-slice-resolution reconstruction. λTik is the Tikhonov regularization parameter. It was set to 0.2 to achieve a high SNR gain and sharp slice resolution (9). We note that the A matrix employed in our reconstructions did not include slice profile information, but this could be flexibly incorporated (10) into Eq1.
Results
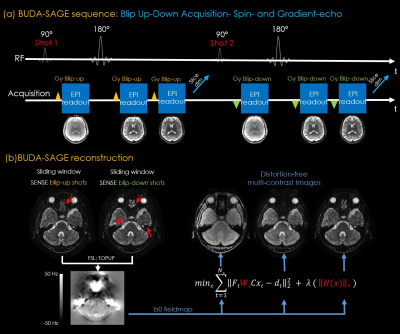
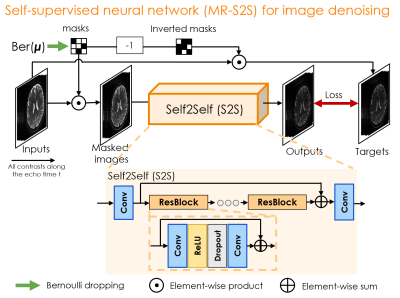
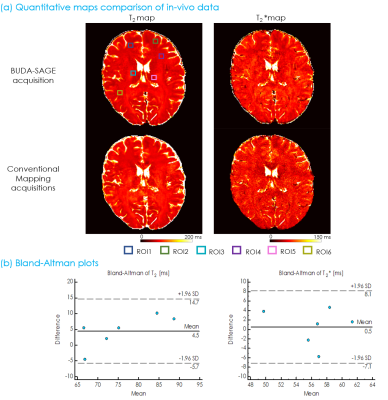
Figure 1 displays the BUDA-SAGE sequence and reconstruction pipeline. The network architecture of self-supervised MR-S2S model for image denoising are provided in Figure 2. T2/T2* maps were obtained using Bloch dictionary matching using the multi-contrast images. Reference T2/T2* maps were calculated by performing a voxel-wise mono-exponential fit to the echo images. Six ROIs were defined as 9×9 voxel boxes throughout the displayed slice, and the results from Bland-Altman analysis are shown in Figure 3(b). These ROIs demonstrate consistent values between the proposed and conventional mapping methods and all points were within the limits of agreement, though some slight biases (T2: 4.5ms, T2*: 0.5ms) were observed.Figure 4(b)&(c) shows whole-brain, distortion-free 1mm isotropic resolution Slider super-resolution reconstruction of T2*-, T2’- and T2-weighted images, demonstrating the ability of Slider reconstruction to provide high-resolution data.
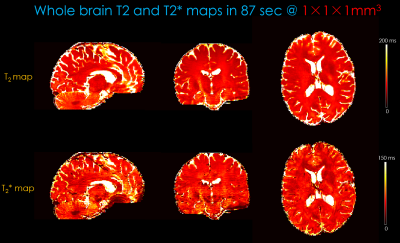
Figure 5 shows whole-brain, distortion-free T2/T2* maps at 1×1×1mm3 resolution using two-groups of 4-shots Slider-encoded(Nslider=2) BUDA-SAGE acquisition from an 85s scan, plus a 2s calibration scan for coil sensitivities.
Conclusion
In this work, BUDA-SAGE with self-supervised denoising and Slider encoding was proposed for distortion-free multi-contrast and quantitative imaging with the ability to provide whole-brain coverage at 1mm isotropic resolution under 90 seconds. This provides a favorable balance between acquisition speed, image quality and the accuracy of T2/T2* estimation.Acknowledgements
This work was supported in part by National Key R&D Program of China (No: 2020AAA0109502), by the National Natural Science Foundation of China (No: U1809204, 61525106, 61427807, 61701436), the Fundamental Research Funds for the Central Universities, and by NIH grants R01 EB028797, R03 EB031175, U01 EB025162, P41 EB030006, U01 EB026996 and the NVidia Corporation for computing support.
References
1. Bültmann E, Spineli LM, Göhner F, Hartmann H, Lanfermann H. Age-related T2 relaxation times at 3 Tesla as a biomarker of infratentorial brain maturation. Child’s Nervous System 2018;34 doi: 10.1007/s00381-017-3561-4.
2. Cao X, Ye H, Liao C, Li Q, He H, Zhong J. Fast 3D brain MR fingerprinting based on multi‐axis spiral projection trajectory. Magnetic Resonance in Medicine 2019;82 doi: 10.1002/mrm.27726.
3. Ma D, Jones SE, Deshmane A, et al. Development of high-resolution 3D MR fingerprinting for detection and characterization of epileptic lesions. Journal of Magnetic Resonance Imaging 2019;49 doi: 10.1002/jmri.26319.
4. Ma S, Wang N, Fan Z, et al. Three‐dimensional whole‐brain simultaneous T1, T2, and T1 ρ quantification using MR Multitasking: Method and initial clinical experience in tissue characterization of multiple sclerosis. Magnetic Resonance in Medicine 2021;85 doi: 10.1002/mrm.28553.
5. Wang F, Dong Z, Reese TG, Rosen B, Wald LL, Setsompop K. 3D Echo Planar Time-resolved Imaging (3D-EPTI) for ultrafast multi-parametric quantitative MRI. doi: 10.1101/2021.05.06.443040.
6. Zhang Z, Wang L, Cho J, et al. BUDA-SAGE with self-supervised denoising enables fast, distortion-free, high-resolution T2, T2*, para- and dia-magnetic susceptibility mapping. 2021.
7. Greenspan H, Oz G, Kiryati N, Peled S. MRI inter-slice reconstruction using super-resolution. Magnetic Resonance Imaging 2002;20 doi: 10.1016/S0730-725X(02)00511-8.
8. Vu AT, Beckett A, Setsompop K, Feinberg DA. Evaluation of SLIce Dithered Enhanced Resolution Simultaneous MultiSlice (SLIDER-SMS) for human fMRI. NeuroImage 2018;164 doi: 10.1016/j.neuroimage.2017.02.001.
9. Cao X, Wang K, Liao C, et al. Efficient T 2 mapping with blip‐up/down EPI and gSlider‐SMS (T 2 ‐BUDA‐gSlider). Magnetic Resonance in Medicine 2021;86 doi: 10.1002/mrm.28872.
10. Van AT, Aksoy M, Holdsworth SJ, Kopeinigg D, Vos SB, Bammer R. Slab profile encoding (PEN) for minimizing slab boundary artifact in three-dimensional diffusion-weighted multislab acquisition. Magnetic Resonance in Medicine 2015;73 doi: 10.1002/mrm.25169.
Figures