3808
Open-Source Hypothalamic-ForniX (OSHy-X) Atlases and Segmentation Tool for 3T and 7T1School of Biomedical Sciences, The University of Queensland, St Lucia, Australia, 2Department of Neurology, Royal Brisbane and Women's Hospital, Brisbane, Australia, 3Wesley Medical Research, The Wesley Hospital, Brisbane, Australia, 4Centre for Clinical Research, The University of Queensland, Brisbane, Australia, 5Australian Institute of Bioengineering and Nanotechnology, The University of Queensland, St Lucia, Australia, 6QIMR Berghofer Medical Research Institute, Brisbane, Australia, 7School of Information Technology and Electrical Engineering, The University of Queensland, St Lucia, Australia, 8Centre for Advanced Imaging, The University of Queensland, Brisbane, Australia, 9Health and Biosecurity, The Commonwealth Scientific and Industrial Research Organisation (CSIRO), Brisbane, Australia
Synopsis
Segmentation and volumetric analysis of the hypothalamus and fornix plays a critical role in improving the understanding of degenerative processes that might impact the function of these structures. We present Open-Source Hypothalamic-ForniX (OSHy-X) atlases and tool for multi-atlas fusion segmentation for 3T and 7T. The atlases are based on 20 manual segmentations, which we demonstrate have high interrater agreement. The versatility of the OSHy-X tool allows segmentation and volumetric analysis for different field strengths and contrasts. We also demonstrate that OSHy-X segmentation has higher Dice overlaps (3T and 7T inputs: p<0005, p<0.005) than a deep-learning segmentation method for the hypothalamus.
Introduction
Segmentation of small structures of the brain including the hypothalamus and fornix is important for primary research of health and disease. Amyotrophic Lateral Sclerosis (ALS) is a fatal neurodegenerative disease that involves the degeneration and death of motor neurons in the brain and spinal cord. Neuronal death and gross volume loss has also been reported in the hypothalamus in ALS1-3. To measure such changes, methods for in vivo MRI segmentation of the hypothalamus and fornix include deep learning4, seed growing techniques5, and manual segmentation1. There is a need to develop and distribute open-source atlases of these structures for more accurate and standardised segmentation. Here, we present the Open-Source Hypothalamic-ForniX (OSHy-X) atlases and tool for multi-atlas fusion segmentation for 3T and 7T. OSHy-X is an atlas repository and containerised Python script that automatically segments the hypothalamus and fornix at 3T and 7T in both T1w and T2w scans. OSHy-X was validated using 3T and 7T data from patients with ALS and non-neurodegenerative disease study participants.Methods
AtlasMR images were acquired from two studies: Endocrine and Appetite Targets and Therapies for MND (EATT4MND) and the Neurobiology-informed diagnostic toolkit for neurodegenerative diseases (7TEA).
Participants in the EATT4MND study were scanned using a 3T Siemens Prisma scanner, with a 3D 1 mm3 isotropic MP2RAGE sequence6 (TR/TE/TIs/TA=5000ms/2.98ms/701,2500ms/9m:02s) and a 1mm3 isotropic T2w FLuid-Attenuated Inversion Recovery scan (TR/TE/TI/TA=5000ms/386ms/1800ms/5m:52s). Participants in the 7TEA study were scanned using a 7T Siemens Magnetom scanner (Siemens Healthcare, Erlangen), with a 0.9 mm3 isotropic MP2RAGE sequence6 (TR/TE/TIs/TA=4900ms/3.1ms/700,2700ms/5m:54s) and a 0.4x0.4x0.8mm3 2D Turbo Spin-Echo scan (TR/TE/TI/TA=10300ms/102ms/1800ms/4m:12s).
Twenty atlases were derived from manual segmentation of the hypothalamus-fornix, conducted by two tracers familiar with the hypothalamus and fornix. Ten non-neurodegenerative disease participants and ten patients with ALS were selected at random from within the larger datasets of the EATT4MND and 7TEA studies for the tracing. Protocols developed by Bocchetta et al. 20157 and Amaral et al. 20188 were used to segment the hypothalamus and fornix, respectively.
Tool
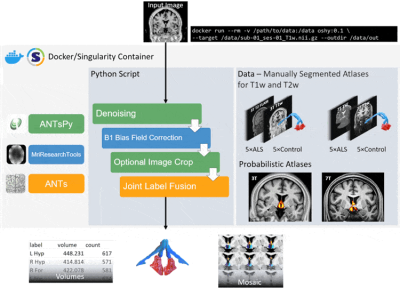
A summary of the pipeline is illustrated in Figure 1. The user can specify the contrast (T1w/T2w) of the atlases used, the field strength (3T/7T) and any pre-processing steps. OSHy-X utilises Joint Label Fusion (JLF)9 from Advanced Normalization Tools (ANTs; v2.3.1) for the registration10 of atlases and segmentation of the target image9. B1+ bias field inhomogeneity correction is performed using MriResearchTools (v0.5.2). Denoising and cropping are performed using ANTs in Python (ANTsPy; v0.2.0).
Performance
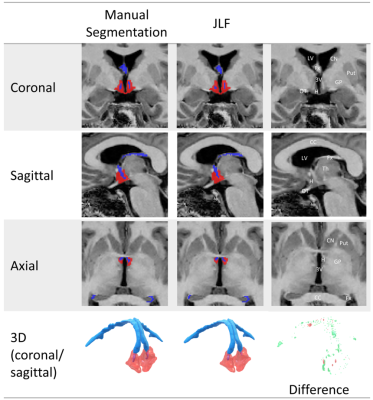
We examined the performance of the JLF method using Dice overlaps of manual segmentations of the hypothalamus-fornix (landmarks used to inform borders of the hypothalamus-fornix are illustrated in Figure 2). We used a leave-one out cross validation (LOOCV) method, whereby each of the atlases used in the JLF were sequentially removed before they were segmented, to ensure fairness in comparisons. We measured the consistency of our raters’ measurements using Dice overlap and intraclass correlation (ICC; 2-way fixed-rater mixed effects model with single measurement). Finally, to gauge the performance of our tool, we compared the Dice overlaps of the manual segmentation to two methods: 1) the OSHy-X tool and 2) a deep learning method for segmenting the hypothalamus4 (available in FreeSurfer v7.2).
Results
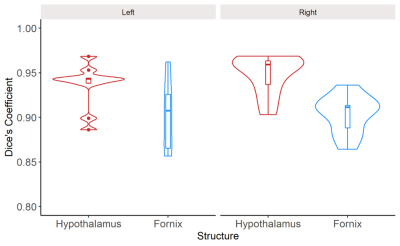
Figure 2 visually compares the differences in the segmentation of a representative non-neurodegenerative disease participant using manual segmentation and JLF using LOOCV. Overall, JLF tends to under-segment throughout the hypothalamus and fornix. To a lesser extent, JLF tends to over-segment the anterior and lateral hypothalamus and the body of the fornix.Dice overlaps (Figure 3) and ICC between the two raters indicate excellent segmentation accuracy. The left and right hypothalamus received scores of 0.90 (0.66-0.98 CI) and 0.91 (0.68-0.98 CI). The left and right fornix received scores of 0.97 (0.87-0.99 CI) and 0.68 (0.13-0.91 CI).
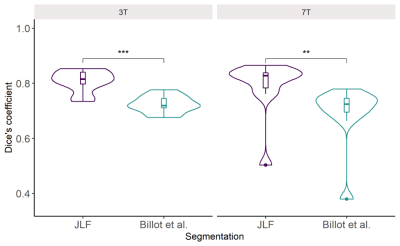
In comparison to a deep learning method for segmentation4, we found that JLF has higher Dice overlaps with the manual segmentations for both 3T and 7T (Figure 4). Additionally, we found that compared to cropped priors, whole-brain priors for JLF offers modest benefits to segmentation accuracy at 3T, but significant performance benefits at 7T compared to the deep learning method. While whole brain instead of cropped priors for JLF improves segmentation performance, computational time increases prohibitively.
Discussion and conclusion
We present an open-source hypothalamus and fornix atlas and segmentation tool for segmenting the hypothalamus-fornix. This atlas is freely available at (https://osf.io/zge9t) and the tool is available via the Neurodesk data analysis environment (https://neurodesk.github.io) or as a Docker/Singularity container (https://github.com/Cadaei-Yuvxvs/OSHy-X). The atlas contains 20 manually labelled scans of both the hypothalamus and fornix at both 3T and 7T field strengths and at multiple contrasts; and contains imaging data for both non-neurodegenerative disease participants and patients with ALS.The segmentation tool can be run on any operating system supporting Docker, requiring a T1w or T2w scan as input. The tool outputs hypothalamus/fornix segmentations and is specifically designed to accommodate for different field strengths, contrasts, and patient populations, though with best performance attained with longer runtimes (whole-brain priors). We contend that “cutting corners” in image segmentation to reduce runtime may compromise the quality of segmentation.
Acknowledgements
We thank all individuals who took part in the studies.
Funding for EATT4MND was provided by Wesley Medical Research (The Wesley Hospital, Brisbane) and The Faculty of Medicine, The University of Queensland.
The authors acknowledge the facilities and scientific and technical assistance of the National Imaging Facility, a National Collaborative Research Infrastructure Strategy (NCRIS) capability, at the Centre for Advanced Imaging, The University of Queensland. This research was undertaken with the assistance of resources and services from the Queensland Cyber Infrastructure Foundation (QCIF). The authors gratefully acknowledge Aiman Al Najjer, Nicole Atcheson, Anita Burns, Saskia Bollmann, and Amelia Ceslis for acquiring data.
JC is supported by the UQ Graduate School Scholarship (RTP). STN is supported by a FightMND Mid-Career Fellowship. MB acknowledges funding from Australian Research Council Future Fellowship grant FT140100865. MB and SB acknowledge the ARC Training Centre for Innovation in Biomedical Imaging Technology (CIBIT). TS is supported by a Motor Neurone Disease Research Australia (MNDRA) Postdoctoral Research Fellowship (PDF2112).References
1. Gorges M, Vercruysse P, Müller H P, et al. Hypothalamic atrophy is related to body mass index and age at onset in amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2017;88(12):1033-1041.
2. Gabery S, Ahmed R M, Caga J, et al. Loss of the metabolism and sleep regulating neuronal populations expressing orexin and oxytocin in the hypothalamus in amyotrophic lateral sclerosis. Neuropathol Appl Neurobiol. 2021
3. Christidi F, Karavasilis E, Rentzos M, et al. Hippocampal pathology in amyotrophic lateral sclerosis: selective vulnerability of subfields and their associated projections. Neurobiol Aging. 2019;84:178-188.
4. Billot B, Bocchetta M, Todd E, et al. Automated segmentation of the hypothalamus and associated subunits in brain MRI. Neuroimage. 2020;223:117287.
5. Wolff J, Schindler S, Lucas C, et al. A semi-automated algorithm for hypothalamus volumetry in 3 Tesla magnetic resonance images. Psychiatry Res Neuroimaging. 2018;277:45-51.
6. Marques J P, Kober T, Krueger G, et al. MP2RAGE, a self bias-field corrected sequence for improved segmentation and T1-mapping at high field. Neuroimage. 2010;49(2):1271-81.
7. Bocchetta M, Gordon E, Manning E, et al. Detailed volumetric analysis of the hypothalamus in behavioral variant frontotemporal dementia. J Neurol. 2015;262(12):2635-42.
8. Amaral R S C, Park M T M, Devenyi G A, et al. Manual segmentation of the fornix, fimbria, and alveus on high-resolution 3T MRI: Application via fully-automated mapping of the human memory circuit white and grey matter in healthy and pathological aging. Neuroimage. 2018;170:132-150.
9. Wang H, Suh J W, Das S R, et al. Multi-Atlas Segmentation with Joint Label Fusion. IEEE Trans Pattern Anal Mach Intell. 2013;35(3):611-23.
10. Avants B B, Epstein C L, Grossman M, et al. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 2008;12(1):26-41.
Figures