3798
Lesion Segmentation for Venous Malformations Based on Unet++ Architecture1Department of Medical Imaging, Henan Provincial People's Hospital, Zhengzhou, China
Synopsis
In this study, we trained a fully automatic lesion segmentation model of venous malformations based on Unet++ structure and fat-saturated T2-weighted images. The results of automatic segmentation of lesions in different sequence directions and locations of the lesions are consistent with the results of manual segmentation by radiologists. The Dice coefficient on the test set reached 0.847. This fully automatic lesion segmentation model can provide support for subsequent automatic diagnosis studies related to venous malformations.
Introduction
Venous malformations (VMs) are slow-flow vascular malformations that are composed of interconnected ectatic venous channels deficient in vascular smooth muscle [1, 2]. VMs are congenital malformations and always present at birth and grow proportionately to the child and expand slowly. Since VMs may occur in any anatomical site or tissue, including head and neck, extremity, and bone, the diagnosis and treatment of venous malformations can be challenging. For venous malformations of the extremity, the lesions can be superficial or deep, focal or diffuse, involving the skin, subcutaneous tissue, musculoskeletal structures and sometimes bone. The differential diagnosis of VMs includes deep infantile hemangioma, capillary malformations (CM), lymphatic malformations (LM), arteriovenous malformations (AVM), and other combined vascular malformations. Magnetic resonance imaging (MRI) represents the gold standard for VMs diagnosis, which can quantify the extent of malformation and the degree of involvement of nearby tissues [3]. Commonly used magnetic resonance sequences include T1-weighted sequence and fat-saturated T2-weighted sequence, which are fat-sensitive and fluid-sensitive sequences respectively.Radiomics represents a method of extracting large amounts of high-dimensional quantitative features from multimodal medical images. Nowadays, more and more studies demonstrate automatic diagnosis models based on radiomics features and machine learning classifiers can be used as an effective tool to improve disease diagnosis and management [4]. In radiomics, lesion segmentation is a fundamental step that determine the regions of feature extraction. The segmentation methods can be manual, semiautomatic, and automatic. There are some shortcomings for manual segmentation of venous malformations. On the one hand, manual segmentation is very consuming. For large focal or diffuse lesions of VMs, radiologists need to delineate the region of interest on dozens of slices which is not suitable for processing large sample data. On the other hand, the inter- and intra-reader variability of interpretation of lesion boundary may be large due to irregular and complex contours, which influence the stability of extracted radiomics features.
In our study, we trained a fully automatic venous malformation lesion segmentation model based on the Unet++ [5] structure, which is suitable for axial, coronal, and sagittal fat-saturated T2-weighted images.
Methods
This study included 172 venous malformation patients who underwent magnetic resonance imaging (MRI) including fat-saturated T2-weighted imaging. The acquisition equipment includes three different models including MAGNETOM Verio 3.0T, TrioTim 3.0T, and Prisma 3.0T MR scanner (Siemens Healthcare, Erlangen, Germany). A total of 537 fat-saturated T2WI sequences were used to train and test the segmentation model, including 257 axial sequences and 280 coronal/sagittal sequences and the test set included 50 axial and 50 coronal/sagittal sequences. The locations of these lesions include maxillofacial, neck, shoulders, chest wall, hands, forearms, arms, thighs, knees, calves, and feet, and include focal and diffuse lesions in morphology. All VMs lesions were segmented by one radiologist using ITK-SNAP tool (www.itksnap.org) and saved as mask files. Then the original T2 weighted images and corresponding mask files were converted to single-slice two-dimensional PNG images. The final training set includes 9087 slice images and corresponding mask files, and the test set includes 2012 slice images and corresponding mask files.In our study, the segmentation model was constructed based on Unet++ architecture which was a powerful architecture for medical image segmentation. Compared with the original Unet architecture, the Unet++ achieved higher on multiple medical image segmentation taks including liver and nodule segmentation on CT images, nuclei segmentation in the microscopy images, etc [5]. The VMs segmentation model was constructed based on Python 3.8.5 and Python libraries including Pytorch, segmentation-models-pytorch, albumentations, and MONAI. The image augmentation methods were applied to enhance the generalization of model and prevent over-fitting. The augmentation methods include horizontal flip, shift, scale, rotate, randomly crop, blur, randomly change brightness and contrast, contrast limited adaptive histogram equalization (CLAHE), etc. The Intersection-over-Union (IoU) and Dice score were used as metrics of model segmentation performance.
Results
During 100 iterations of training and validation, the lesion segmentation model achieved the best performance at 80th iteration with a learning rate of 10-5. The performance of the best model achieved Dice 0.847 and IoU 0.744 in the training set and Dice 0.807 and IoU 0.749 in the test set.The lesion areas outlined by the radiologist and predicted areas of the segmentation model were shown in Figure 1-2, which contains different lesion locations (maxillofacial, chest wall, forearm, hand, thigh, calf) and sequence directions (axial, coronal, sagittal). Compared with manual segmentation, the automatic segmentation model has consistent results, and the edges are more refined. The segmentation model can also distinguish hyperintense lesions of VMs from other hyperintense tissues or artifact areas.
Discussion and Conclusions
In manual segmentation, radiologists often use polylines to outline arc-shaped lesion boundaries, which makes boundaries slightly different from actual lesion contours. In comparison, the automatic VMs lesion segmentation model based on deep learning depicts the boundary more finely, which is closer to the actual contour of the lesion.In conclusion, based on the UNet++ architecture and fat-saturated T2 weighted images, we constructed an effective automatic lesion segmentation model for venous malformations, which is suitable for different lesion locations and image directions. The fully automatic lesion segmentation model can provide strong support for subsequent automatic diagnosis of VMs based on massive medical images.
Acknowledgements
No acknowledgement found.References
[1] O. Enjolras, D. Ciabrini, E. Mazoyer, C. Laurian, D. Herbreteau, Extensive pure venous malformations in the upper or lower limb: a review of 27 cases, Journal of the American Academy of Dermatology 36(2 Pt 1) (1997) 219-25.
[2] A.B. Johnson, G.T. Richter, Surgical Considerations in Vascular Malformations, Tech Vasc Interv Radiol 22(4) (2019) 100635.
[3] G. Colletti, A.M. Ierardi, Understanding venous malformations of the head and neck: a comprehensive insight, Medical oncology (Northwood, London, England) 34(3) (2017) 42.
[4] Z. Liu, S. Wang, D. Dong, J. Wei, C. Fang, X. Zhou, K. Sun, L. Li, B. Li, M. Wang, J. Tian, The Applications of Radiomics in Precision Diagnosis and Treatment of Oncology: Opportunities and Challenges, Theranostics 9(5) (2019) 1303-1322.
[5] Z. Zhou, M.M. Rahman Siddiquee, N. Tajbakhsh, J. Liang, UNet++: A Nested U-Net Architecture for Medical Image Segmentation, Springer International Publishing, Cham, 2018, pp. 3-11.
Figures
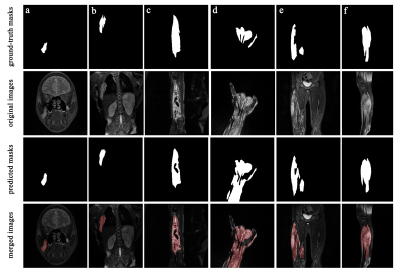
Figure 1. The Original coronal/sagittal fat-saturated T2WI images, ground-truth masks and predicted masks of VMs lesions. Different lesion locations include (a) maxillofacial, (b) chest wall, (c) forearm, (d) hand, (e) thigh, (f) calf. In merged images, the lesion areas segmented by the model were marked in red.
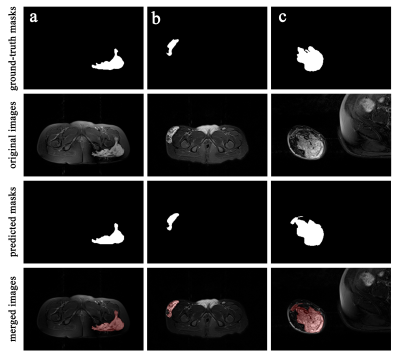
Figure 2. The Original axial fat-saturated T2WI images, ground-truth masks and predicted masks of VMs lesions. In merged images, the lesion areas segmented by the model were marked in red.